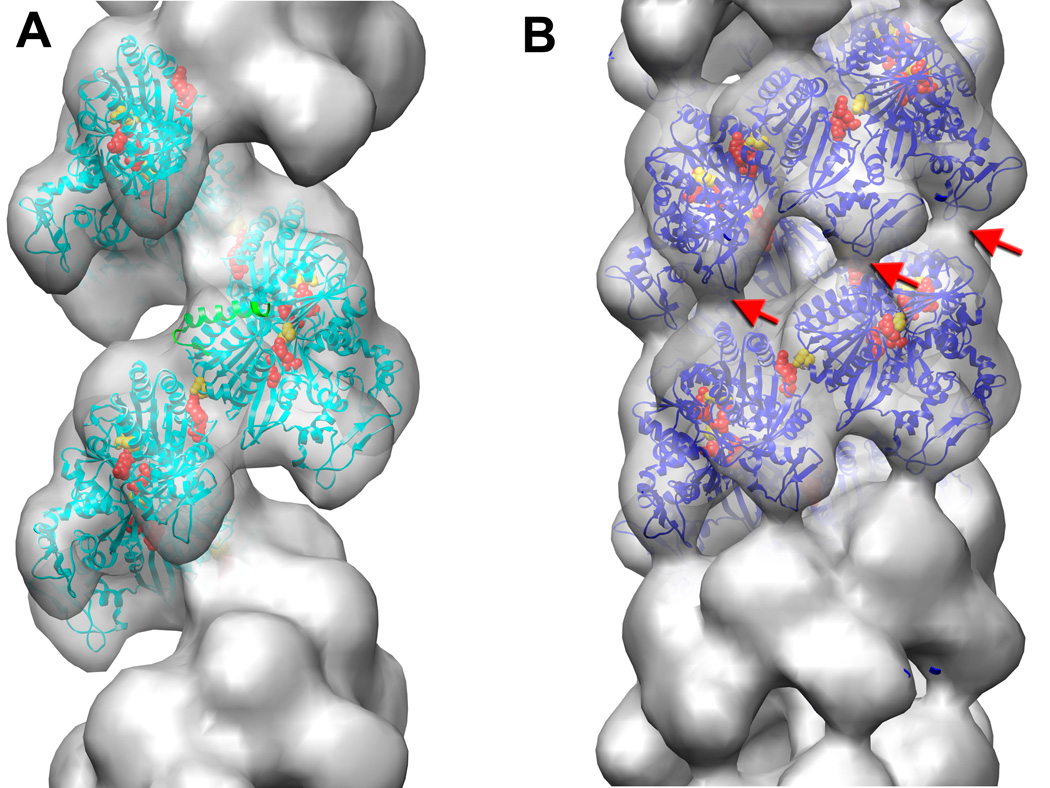
Fig. 4.
Electron microscopy of UvsX recombination filaments. (A) Reconstruction of the extended ‘active’ filament (grey) formed in the presence of dsDNA and ATP into which the UvsX crystal structure has been fitted (cyan). The C-terminal helical domain is pointing down towards the large groove. The filament has a rotation per subunit of 58.5° and axial rise per subunit of 16.1 Å. The 28 N-terminal residues of RecA were used to model the missing N-terminal UvsX residues (green ribbons). The positions of three residues in UvsX at the monomer-monomer interface that correspond to those in RecA involved in the ATP hydrolysis are shown as red (Lys246, Arg248), and yellow (Glu92) spheres. (B) The compressed ‘inactive’ filament formed in the presence of dsDNA and ADP in which the fitted UvsX structure is shown in dark blue. The filament has a rotation per subunit of 55.7° and axial rise per subunit of 10.8 Å. A bridge of density across the groove, corresponding to an interaction between residues 130–132 of one monomer and residues 285–288 of the other monomer, is indicated by red arrows.