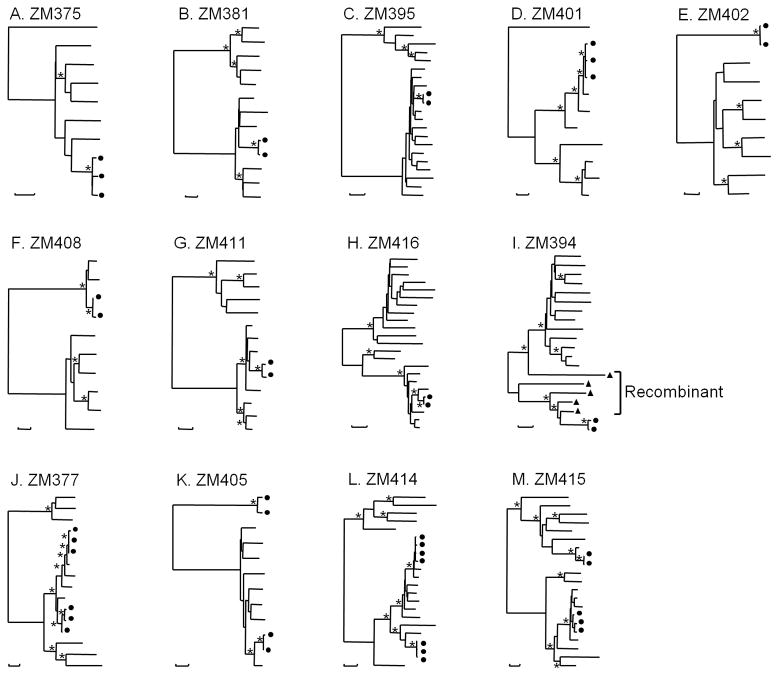
Figure 2.
Phylogenetic tree analysis of clonal expansion env gene sequences. Midpoint-rooted phylogenetic trees were constructed with all env sequences from each individual who harbored clonal expansion viruses using the neighbor-joining method and the Kimura two-parameter model. Horizontal branch lengths are drawn to scale (the scale bar represents 0.005 nucleotide substitutions per site); vertical separation is for clarity only. Asterisks indicate bootstrap values in which the cluster to the right is supported in 85% or more replicates (out of 1,000). Patterns of identity or near identity are marked with black dots at the terminal leaves.