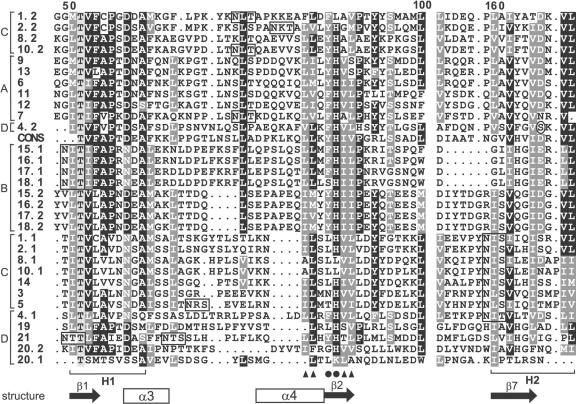
Figure 1.
Sequence alignment of part of the fascilcin domains of the Arabidopsis FLAs (1-21) and a consensus sequence (CONS) generated from 78 fasciclin domains from a diverse range of species. Where there are two fasciclin domains in the protein, the domain closest to the N terminus is indicated by .1 and the second by .2. The alignment was generated by ClustalX and manually edited. Conserved and identical residues are shaded dark gray and black, respectively, and similar residues are indicated in light gray. Periods represent gaps introduced by the program for optimal alignment, and the space before the H2 domain corresponds to 52 amino acid residues (or gaps) that show little or no similarity. The H1 and H2 conserved regions characteristic of fasciclin domains are indicated below the alignment. Amino acids thought to be involved in adhesion (Ile, Leu, and Val; Kim et al., 2000, 2002) are indicated by black triangles and YH motifs by black circles. The predicted secondary structure elements of the fasciclin domain based on the crystal structure of fas3 and fas4 domains of fruitfly are indicated below the alignment. These secondary structure elements are in most FLAs, but some, e.g. α4, are not conserved in group B FLAs (see “Discussion”). N-glycosylation motifs are boxed. The amino acid in the H2 region of FLA4 that is substituted in the sos5 mutant (Ser to Phe) is circled. The FLAs are grouped into four groups (A-D) based on phylogenetic analysis and pair-wise sequence comparison (see “Results”), as indicated on the left-hand side of the alignment.