Abstract
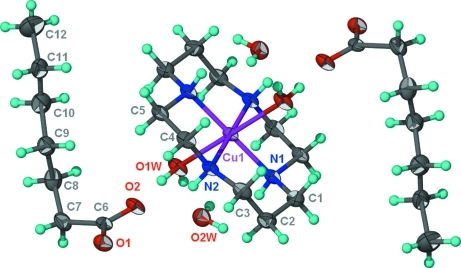
The CuII atom in the title salt, [Cu(C10H24N4)(H2O)2][CH3(CH2)5CO2]2·2H2O, is chelated by the four N atoms of the 1,4,8,11-tetraazacyclotetradecane (cyclam) ligand and is coordinated by two water molecules in a tetragonally Jahn–Teller-distorted octahedral geometry. The CuII atom lies on a center of inversion. The cations, anions and uncoordinated water molecules are linked by N—H⋯O and O—H⋯O hydrogen bonds, forming a layer structure parallel to (100). The alkyl chain of the anion is disordered over two positions in a 0.82 (1):0.18 (1) ratio.
Related literature
For related diaqua(1,4,8,11-tetraazacyclotetradecane)copper carboxylates, see: Lindoy et al. (2003 ▶); Hunter et al. (2005 ▶).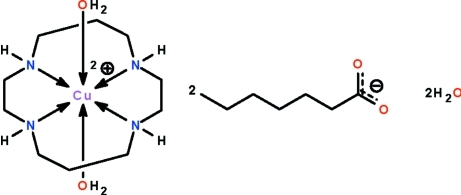
Experimental
Crystal data
[Cu(C10H24N4)(H2O)2](C7H13O2)2·2H2O
M r = 594.28
Monoclinic,
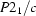
a = 11.7257 (6) Å
b = 9.9426 (5) Å
c = 13.4573 (7) Å
β = 103.1363 (7)°
V = 1527.85 (14) Å3
Z = 2
Mo Kα radiation
μ = 0.76 mm−1
T = 100 K
0.35 × 0.25 × 0.15 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.776, T max = 0.894
14358 measured reflections
3506 independent reflections
3140 reflections with I > 2σ(I)
R int = 0.022
Refinement
R[F 2 > 2σ(F 2)] = 0.028
wR(F 2) = 0.079
S = 1.06
3506 reflections
212 parameters
12 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.81 e Å−3
Δρmin = −0.31 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810025687/bt5285sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810025687/bt5285Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Cu1—N1 | 2.026 (1) |
| Cu1—N2 | 2.025 (1) |
| Cu1—O1w | 2.499 (1) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.86 (1) | 2.30 (1) | 2.983 (2) | 137 (2) |
| N2—H2⋯O2 | 0.86 (1) | 2.12 (1) | 2.924 (2) | 156 (2) |
| O1w—H11⋯O2 | 0.83 (1) | 1.93 (1) | 2.730 (2) | 162 (2) |
| O1w—H12⋯O2w | 0.83 (1) | 1.95 (1) | 2.777 (2) | 174 (2) |
| O2w—H21⋯O1ii | 0.83 (1) | 2.02 (1) | 2.833 (2) | 166 (2) |
| O2w—H22⋯O1i | 0.84 (1) | 1.91 (1) | 2.743 (2) | 171 (2) |
Symmetry codes: (i) 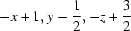 ; (ii)
; (ii) 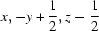 .
.
Acknowledgments
We thank the University of Malaya (RG039/09SUS) and the Ministry of Higher Education (FP017/2009) for supporting this study.
supplementary crystallographic information
Comment
The copper(II) ion forms a number of complexes with 1,4,8,11-tetraazacyclotetradecane in which the metal atom is coordinated by the four amino donor-atoms of the cyclic ligand. Among the carboxylate derivatives, neither the acetate nor the benzoate ions bind directly with the copper atom. The copper atom is coordinated instead by water molecules so that the carboxylate group interacts indirectly with the metal atom through the coordinated water molecules (Hunter et al., 2005; Lindoy et al., 2003). The copper(II) atom in the salt, [Cu(H2O)2(C10H24N4)]2+ 2[CH3(CH2)5CO2]-.2H2O (Scheme I), is chelated by the four nitrogen atoms of the cyclam ligand and is coordinated by two water molecules in a Jahn-Teller type of tetragonally distorted octahedral geometry. The copper atom lies on a center of inversion (Fig. 1). The cations, anions and lattice water molecules are linked by N–H···O and O–H···O hydrogen bonds to form a layer structure.
Experimental
1,4,8,11-Tetraazacyclotetradecane (0.50 g, 2.50 mmol) dissolved in ethanol (25 ml) was mixed with a suspension of copper heptanoate (0.80 g, 2.5 mmol) in ethanol (50 ml) to give a purple solution. The solution was heated for an hour and then filtered. Prismatic crystals separated from the solution when it was left to cool slowly.
Refinement
Carbon-bound H-atoms were placed in calculated positions (C—H 0.95 to 0.99 Å) and were included in the refinement in the riding model approximation, with U(H) set to 1.2 to 1.5U(C).
The amino and water H-atoms were located in a difference Fourier map, and were refined with distance restraints of N–H···O 0.86±0.01, O–H 0.84±0.01 Å; their isotropic displacement parameters were freely refined.
The alkyl chain of the carboxylate ion is disordered over two positions; the disorder refined to an 82 (1):18 (1) ratio. Bond distances for each pair of bonds were restrained to within 0.01Å of each other. The displacement parameters of the primed atoms were constrained to be equal of those of the unprimed ones.
Figures
Fig. 1.
Anisotropic displacement ellipsoid plot (Barbour, 2001) of [Cu(H2O)2(C10H24N4)]2+ 2[CH3(CH2)5CO2]-.2H2O at the 70% probability level; hydrogen atoms are drawn as spheres of arbitrary radius. The disorder in the alkyl chain is not shown.
Crystal data
| [Cu(C10H24N4)(H2O)2](C7H13O2)2·2H2O | F(000) = 646 |
| Mr = 594.28 | Dx = 1.292 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 8003 reflections |
| a = 11.7257 (6) Å | θ = 2.6–28.3° |
| b = 9.9426 (5) Å | µ = 0.76 mm−1 |
| c = 13.4573 (7) Å | T = 100 K |
| β = 103.1363 (7)° | Block, purple |
| V = 1527.85 (14) Å3 | 0.35 × 0.25 × 0.15 mm |
| Z = 2 |
Data collection
| Bruker SMART APEX diffractometer | 3506 independent reflections |
| Radiation source: fine-focus sealed tube | 3140 reflections with I > 2σ(I) |
| graphite | Rint = 0.022 |
| ω scans | θmax = 27.5°, θmin = 1.8° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −15→15 |
| Tmin = 0.776, Tmax = 0.894 | k = −12→12 |
| 14358 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.028 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.079 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0406P)2 + 0.7328P] where P = (Fo2 + 2Fc2)/3 |
| 3506 reflections | (Δ/σ)max = 0.001 |
| 212 parameters | Δρmax = 0.81 e Å−3 |
| 12 restraints | Δρmin = −0.31 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cu1 | 0.5000 | 0.5000 | 0.5000 | 0.01672 (8) | |
| O1 | 0.37230 (9) | 0.57029 (10) | 0.88828 (7) | 0.0254 (2) | |
| O2 | 0.35970 (10) | 0.48007 (10) | 0.73553 (8) | 0.0260 (2) | |
| O1W | 0.35364 (10) | 0.35174 (11) | 0.55509 (8) | 0.0262 (2) | |
| H11 | 0.3445 (19) | 0.378 (2) | 0.6109 (10) | 0.047 (6)* | |
| H12 | 0.3586 (17) | 0.2684 (10) | 0.5589 (15) | 0.035 (5)* | |
| O2W | 0.38768 (10) | 0.07550 (11) | 0.57173 (8) | 0.0272 (2) | |
| H21 | 0.372 (2) | 0.031 (2) | 0.5184 (11) | 0.045 (6)* | |
| H22 | 0.4606 (9) | 0.070 (2) | 0.5895 (16) | 0.046 (6)* | |
| N1 | 0.62254 (10) | 0.35391 (11) | 0.53855 (9) | 0.0196 (2) | |
| H1 | 0.5925 (14) | 0.2927 (14) | 0.5699 (12) | 0.025 (4)* | |
| N2 | 0.54008 (11) | 0.58808 (11) | 0.63930 (8) | 0.0196 (2) | |
| H2 | 0.5052 (14) | 0.5435 (16) | 0.6783 (11) | 0.022 (4)* | |
| C1 | 0.73634 (13) | 0.39280 (15) | 0.60465 (11) | 0.0246 (3) | |
| H1A | 0.7760 | 0.4571 | 0.5676 | 0.030* | |
| H1B | 0.7866 | 0.3120 | 0.6205 | 0.030* | |
| C2 | 0.72217 (14) | 0.45693 (16) | 0.70380 (11) | 0.0267 (3) | |
| H2A | 0.8003 | 0.4645 | 0.7507 | 0.032* | |
| H2B | 0.6740 | 0.3967 | 0.7363 | 0.032* | |
| C3 | 0.66565 (13) | 0.59536 (15) | 0.69082 (10) | 0.0242 (3) | |
| H3A | 0.6738 | 0.6376 | 0.7587 | 0.029* | |
| H3B | 0.7070 | 0.6528 | 0.6502 | 0.029* | |
| C4 | 0.48410 (14) | 0.72262 (14) | 0.62714 (11) | 0.0244 (3) | |
| H4A | 0.5314 | 0.7860 | 0.5964 | 0.029* | |
| H4B | 0.4781 | 0.7584 | 0.6944 | 0.029* | |
| C5 | 0.36356 (13) | 0.70700 (14) | 0.55851 (10) | 0.0231 (3) | |
| H5A | 0.3146 | 0.6484 | 0.5914 | 0.028* | |
| H5B | 0.3249 | 0.7958 | 0.5453 | 0.028* | |
| C6 | 0.31775 (13) | 0.50923 (13) | 0.80991 (11) | 0.0203 (3) | |
| C7 | 0.18993 (17) | 0.4746 (2) | 0.81018 (17) | 0.0289 (5) | 0.822 (3) |
| H7A | 0.1856 | 0.3788 | 0.8288 | 0.035* | 0.822 (3) |
| H7B | 0.1655 | 0.5291 | 0.8634 | 0.035* | 0.822 (3) |
| C8 | 0.10391 (17) | 0.49936 (19) | 0.70811 (18) | 0.0318 (5) | 0.822 (3) |
| H8A | 0.1308 | 0.4489 | 0.6542 | 0.038* | 0.822 (3) |
| H8B | 0.0263 | 0.4633 | 0.7119 | 0.038* | 0.822 (3) |
| C9 | 0.09044 (18) | 0.6455 (2) | 0.67784 (15) | 0.0299 (5) | 0.822 (3) |
| H9A | 0.1691 | 0.6840 | 0.6812 | 0.036* | 0.822 (3) |
| H9B | 0.0556 | 0.6942 | 0.7279 | 0.036* | 0.822 (3) |
| C10 | 0.01464 (18) | 0.6690 (2) | 0.57143 (18) | 0.0357 (5) | 0.822 (3) |
| H10A | 0.0433 | 0.6106 | 0.5227 | 0.043* | 0.822 (3) |
| H10B | −0.0665 | 0.6411 | 0.5709 | 0.043* | 0.822 (3) |
| C11 | 0.01253 (18) | 0.8120 (3) | 0.53458 (18) | 0.0402 (6) | 0.822 (3) |
| H11A | 0.0928 | 0.8384 | 0.5306 | 0.048* | 0.822 (3) |
| H11B | −0.0118 | 0.8714 | 0.5851 | 0.048* | 0.822 (3) |
| C12 | −0.0696 (3) | 0.8339 (5) | 0.4308 (2) | 0.0541 (9) | 0.822 (3) |
| H12A | −0.0671 | 0.9286 | 0.4112 | 0.081* | 0.822 (3) |
| H12B | −0.1497 | 0.8101 | 0.4344 | 0.081* | 0.822 (3) |
| H12C | −0.0450 | 0.7771 | 0.3799 | 0.081* | 0.822 (3) |
| C7' | 0.2043 (7) | 0.4295 (9) | 0.7967 (10) | 0.0289 (5) | 0.18 |
| H7'1 | 0.1981 | 0.3868 | 0.8617 | 0.035* | 0.178 (3) |
| H7'2 | 0.1995 | 0.3589 | 0.7441 | 0.035* | 0.178 (3) |
| C8' | 0.1079 (7) | 0.5358 (9) | 0.7631 (8) | 0.0318 (5) | 0.18 |
| H8'1 | 0.0302 | 0.4928 | 0.7548 | 0.038* | 0.178 (3) |
| H8'2 | 0.1152 | 0.6057 | 0.8165 | 0.038* | 0.178 (3) |
| C9' | 0.1167 (9) | 0.6010 (11) | 0.6632 (7) | 0.0299 (5) | 0.18 |
| H9'1 | 0.1238 | 0.5297 | 0.6137 | 0.036* | 0.178 (3) |
| H9'2 | 0.1889 | 0.6561 | 0.6750 | 0.036* | 0.178 (3) |
| C10' | 0.0116 (8) | 0.6905 (12) | 0.6166 (8) | 0.0357 (5) | 0.18 |
| H10C | −0.0619 | 0.6384 | 0.6088 | 0.043* | 0.178 (3) |
| H10D | 0.0080 | 0.7676 | 0.6624 | 0.043* | 0.178 (3) |
| C11' | 0.0239 (9) | 0.7419 (15) | 0.5123 (8) | 0.0402 (6) | 0.18 |
| H11C | 0.0363 | 0.6646 | 0.4696 | 0.048* | 0.178 (3) |
| H11D | 0.0934 | 0.8010 | 0.5215 | 0.048* | 0.178 (3) |
| C12' | −0.0852 (14) | 0.820 (3) | 0.4572 (14) | 0.0541 (9) | 0.18 |
| H12D | −0.0733 | 0.8539 | 0.3918 | 0.081* | 0.178 (3) |
| H12E | −0.0983 | 0.8964 | 0.4996 | 0.081* | 0.178 (3) |
| H12F | −0.1536 | 0.7608 | 0.4450 | 0.081* | 0.178 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.02403 (13) | 0.01387 (12) | 0.01321 (12) | −0.00126 (8) | 0.00619 (9) | −0.00200 (7) |
| O1 | 0.0350 (6) | 0.0224 (5) | 0.0193 (5) | 0.0001 (4) | 0.0071 (4) | −0.0030 (4) |
| O2 | 0.0375 (6) | 0.0262 (5) | 0.0170 (5) | −0.0036 (4) | 0.0114 (4) | −0.0006 (4) |
| O1W | 0.0414 (6) | 0.0197 (5) | 0.0214 (5) | −0.0041 (4) | 0.0155 (5) | −0.0017 (4) |
| O2W | 0.0334 (6) | 0.0254 (5) | 0.0243 (5) | 0.0020 (4) | 0.0098 (5) | −0.0037 (4) |
| N1 | 0.0278 (6) | 0.0160 (5) | 0.0168 (5) | −0.0017 (4) | 0.0087 (5) | 0.0008 (4) |
| N2 | 0.0286 (6) | 0.0164 (5) | 0.0154 (5) | −0.0036 (4) | 0.0086 (5) | −0.0012 (4) |
| C1 | 0.0262 (7) | 0.0244 (7) | 0.0236 (7) | 0.0000 (6) | 0.0062 (6) | 0.0026 (5) |
| C2 | 0.0281 (7) | 0.0309 (7) | 0.0195 (7) | −0.0030 (6) | 0.0023 (6) | 0.0020 (6) |
| C3 | 0.0296 (7) | 0.0261 (7) | 0.0167 (6) | −0.0071 (6) | 0.0048 (5) | −0.0043 (5) |
| C4 | 0.0397 (8) | 0.0157 (6) | 0.0200 (7) | −0.0013 (6) | 0.0114 (6) | −0.0040 (5) |
| C5 | 0.0348 (8) | 0.0175 (6) | 0.0201 (7) | 0.0028 (5) | 0.0129 (6) | 0.0003 (5) |
| C6 | 0.0256 (7) | 0.0181 (6) | 0.0180 (6) | −0.0008 (5) | 0.0067 (5) | 0.0044 (5) |
| C7 | 0.0285 (9) | 0.0325 (12) | 0.0286 (11) | −0.0059 (9) | 0.0126 (7) | 0.0039 (9) |
| C8 | 0.0227 (9) | 0.0334 (11) | 0.0399 (13) | −0.0077 (7) | 0.0084 (9) | −0.0050 (8) |
| C9 | 0.0235 (10) | 0.0395 (13) | 0.0263 (10) | −0.0052 (8) | 0.0048 (7) | −0.0021 (8) |
| C10 | 0.0227 (8) | 0.0492 (12) | 0.0325 (12) | −0.0052 (8) | 0.0008 (9) | −0.0009 (10) |
| C11 | 0.0225 (9) | 0.0668 (17) | 0.0313 (11) | 0.0054 (11) | 0.0064 (8) | 0.0068 (11) |
| C12 | 0.0371 (14) | 0.0795 (19) | 0.039 (2) | 0.0062 (13) | −0.0047 (11) | 0.0093 (15) |
| C7' | 0.0285 (9) | 0.0325 (12) | 0.0286 (11) | −0.0059 (9) | 0.0126 (7) | 0.0039 (9) |
| C8' | 0.0227 (9) | 0.0334 (11) | 0.0399 (13) | −0.0077 (7) | 0.0084 (9) | −0.0050 (8) |
| C9' | 0.0235 (10) | 0.0395 (13) | 0.0263 (10) | −0.0052 (8) | 0.0048 (7) | −0.0021 (8) |
| C10' | 0.0227 (8) | 0.0492 (12) | 0.0325 (12) | −0.0052 (8) | 0.0008 (9) | −0.0009 (10) |
| C11' | 0.0225 (9) | 0.0668 (17) | 0.0313 (11) | 0.0054 (11) | 0.0064 (8) | 0.0068 (11) |
| C12' | 0.0371 (14) | 0.0795 (19) | 0.039 (2) | 0.0062 (13) | −0.0047 (11) | 0.0093 (15) |
Geometric parameters (Å, °)
| Cu1—N1 | 2.026 (1) | C7—H7B | 0.9900 |
| Cu1—N2 | 2.025 (1) | C8—C9 | 1.507 (3) |
| Cu1—N2i | 2.025 (1) | C8—H8A | 0.9900 |
| Cu1—N1i | 2.026 (1) | C8—H8B | 0.9900 |
| Cu1—O1w | 2.499 (1) | C9—C10 | 1.522 (3) |
| O1—C6 | 1.2584 (18) | C9—H9A | 0.9900 |
| O2—C6 | 1.2456 (18) | C9—H9B | 0.9900 |
| O1W—H11 | 0.826 (10) | C10—C11 | 1.504 (3) |
| O1W—H12 | 0.831 (9) | C10—H10A | 0.9900 |
| O2W—H21 | 0.829 (10) | C10—H10B | 0.9900 |
| O2W—H22 | 0.836 (10) | C11—C12 | 1.521 (3) |
| N1—C1 | 1.4771 (19) | C11—H11A | 0.9900 |
| N1—C5i | 1.4818 (17) | C11—H11B | 0.9900 |
| N1—H1 | 0.860 (9) | C12—H12A | 0.9800 |
| N2—C3 | 1.4798 (18) | C12—H12B | 0.9800 |
| N2—C4 | 1.4827 (18) | C12—H12C | 0.9800 |
| N2—H2 | 0.858 (9) | C7'—C8' | 1.539 (9) |
| C1—C2 | 1.521 (2) | C7'—H7'1 | 0.9900 |
| C1—H1A | 0.9900 | C7'—H7'2 | 0.9900 |
| C1—H1B | 0.9900 | C8'—C9' | 1.516 (8) |
| C2—C3 | 1.520 (2) | C8'—H8'1 | 0.9900 |
| C2—H2A | 0.9900 | C8'—H8'2 | 0.9900 |
| C2—H2B | 0.9900 | C9'—C10' | 1.535 (9) |
| C3—H3A | 0.9900 | C9'—H9'1 | 0.9900 |
| C3—H3B | 0.9900 | C9'—H9'2 | 0.9900 |
| C4—C5 | 1.511 (2) | C10'—C11' | 1.531 (9) |
| C4—H4A | 0.9900 | C10'—H10C | 0.9900 |
| C4—H4B | 0.9900 | C10'—H10D | 0.9900 |
| C5—N1i | 1.4818 (17) | C11'—C12' | 1.538 (9) |
| C5—H5A | 0.9900 | C11'—H11C | 0.9900 |
| C5—H5B | 0.9900 | C11'—H11D | 0.9900 |
| C6—C7' | 1.524 (8) | C12'—H12D | 0.9800 |
| C6—C7 | 1.539 (2) | C12'—H12E | 0.9800 |
| C7—C8 | 1.528 (3) | C12'—H12F | 0.9800 |
| C7—H7A | 0.9900 | ||
| N2—Cu1—N2i | 180.0 | H7A—C7—H7B | 107.7 |
| N2—Cu1—N1i | 85.92 (5) | C9—C8—C7 | 113.91 (17) |
| N2i—Cu1—N1i | 94.08 (5) | C9—C8—H8A | 108.8 |
| N2—Cu1—N1 | 94.08 (5) | C7—C8—H8A | 108.8 |
| N2i—Cu1—N1 | 85.92 (5) | C9—C8—H8B | 108.8 |
| N1i—Cu1—N1 | 180.0 | C7—C8—H8B | 108.8 |
| N2—Cu1—O1W | 90.63 (4) | H8A—C8—H8B | 107.7 |
| N2i—Cu1—O1W | 89.37 (4) | C8—C9—C10 | 113.94 (18) |
| N1i—Cu1—O1W | 90.23 (4) | C8—C9—H9A | 108.8 |
| N1—Cu1—O1W | 89.77 (4) | C10—C9—H9A | 108.8 |
| Cu1—O1W—H11 | 108.7 (16) | C8—C9—H9B | 108.8 |
| Cu1—O1W—H12 | 124.3 (14) | C10—C9—H9B | 108.8 |
| H11—O1W—H12 | 106 (2) | H9A—C9—H9B | 107.7 |
| H21—O2W—H22 | 103 (2) | C11—C10—C9 | 114.72 (19) |
| C1—N1—C5i | 111.88 (11) | C11—C10—H10A | 108.6 |
| C1—N1—Cu1 | 117.23 (9) | C9—C10—H10A | 108.6 |
| C5i—N1—Cu1 | 106.32 (8) | C11—C10—H10B | 108.6 |
| C1—N1—H1 | 107.7 (12) | C9—C10—H10B | 108.6 |
| C5i—N1—H1 | 106.4 (12) | H10A—C10—H10B | 107.6 |
| Cu1—N1—H1 | 106.7 (12) | C10—C11—C12 | 113.4 (3) |
| C3—N2—C4 | 112.08 (11) | C10—C11—H11A | 108.9 |
| C3—N2—Cu1 | 116.85 (9) | C12—C11—H11A | 108.9 |
| C4—N2—Cu1 | 106.51 (8) | C10—C11—H11B | 108.9 |
| C3—N2—H2 | 107.5 (12) | C12—C11—H11B | 108.9 |
| C4—N2—H2 | 105.9 (12) | H11A—C11—H11B | 107.7 |
| Cu1—N2—H2 | 107.4 (12) | C11—C12—H12A | 109.5 |
| N1—C1—C2 | 111.97 (12) | C11—C12—H12B | 109.5 |
| N1—C1—H1A | 109.2 | H12A—C12—H12B | 109.5 |
| C2—C1—H1A | 109.2 | C11—C12—H12C | 109.5 |
| N1—C1—H1B | 109.2 | H12A—C12—H12C | 109.5 |
| C2—C1—H1B | 109.2 | H12B—C12—H12C | 109.5 |
| H1A—C1—H1B | 107.9 | C6—C7'—C8' | 103.9 (6) |
| C1—C2—C3 | 114.22 (12) | C6—C7'—H7'1 | 111.0 |
| C1—C2—H2A | 108.7 | C8'—C7'—H7'1 | 111.0 |
| C3—C2—H2A | 108.7 | C6—C7'—H7'2 | 111.0 |
| C1—C2—H2B | 108.7 | C8'—C7'—H7'2 | 111.0 |
| C3—C2—H2B | 108.7 | H7'1—C7'—H7'2 | 109.0 |
| H2A—C2—H2B | 107.6 | C9'—C8'—C7' | 111.2 (8) |
| N2—C3—C2 | 111.77 (11) | C9'—C8'—H8'1 | 109.4 |
| N2—C3—H3A | 109.3 | C7'—C8'—H8'1 | 109.4 |
| C2—C3—H3A | 109.3 | C9'—C8'—H8'2 | 109.4 |
| N2—C3—H3B | 109.3 | C7'—C8'—H8'2 | 109.4 |
| C2—C3—H3B | 109.3 | H8'1—C8'—H8'2 | 108.0 |
| H3A—C3—H3B | 107.9 | C8'—C9'—C10' | 113.6 (8) |
| N2—C4—C5 | 107.69 (11) | C8'—C9'—H9'1 | 108.9 |
| N2—C4—H4A | 110.2 | C10'—C9'—H9'1 | 108.9 |
| C5—C4—H4A | 110.2 | C8'—C9'—H9'2 | 108.9 |
| N2—C4—H4B | 110.2 | C10'—C9'—H9'2 | 108.9 |
| C5—C4—H4B | 110.2 | H9'1—C9'—H9'2 | 107.7 |
| H4A—C4—H4B | 108.5 | C11'—C10'—C9' | 109.5 (8) |
| N1i—C5—C4 | 107.82 (11) | C11'—C10'—H10C | 109.8 |
| N1i—C5—H5A | 110.1 | C9'—C10'—H10C | 109.8 |
| C4—C5—H5A | 110.1 | C11'—C10'—H10D | 109.8 |
| N1i—C5—H5B | 110.1 | C9'—C10'—H10D | 109.8 |
| C4—C5—H5B | 110.1 | H10C—C10'—H10D | 108.2 |
| H5A—C5—H5B | 108.5 | C10'—C11'—C12' | 111.7 (10) |
| O2—C6—O1 | 124.58 (14) | C10'—C11'—H11C | 109.3 |
| O2—C6—C7' | 106.2 (5) | C12'—C11'—H11C | 109.3 |
| O1—C6—C7' | 127.8 (5) | C10'—C11'—H11D | 109.3 |
| O2—C6—C7 | 120.84 (14) | C12'—C11'—H11D | 109.3 |
| O1—C6—C7 | 114.56 (14) | H11C—C11'—H11D | 107.9 |
| C7'—C6—C7 | 19.8 (4) | C11'—C12'—H12D | 109.5 |
| C8—C7—C6 | 113.97 (16) | C11'—C12'—H12E | 109.5 |
| C8—C7—H7A | 108.8 | H12D—C12'—H12E | 109.5 |
| C6—C7—H7A | 108.8 | C11'—C12'—H12F | 109.5 |
| C8—C7—H7B | 108.8 | H12D—C12'—H12F | 109.5 |
| C6—C7—H7B | 108.8 | H12E—C12'—H12F | 109.5 |
| N2—Cu1—N1—C1 | −38.82 (10) | C3—N2—C4—C5 | 170.38 (11) |
| N2i—Cu1—N1—C1 | 141.18 (10) | Cu1—N2—C4—C5 | 41.42 (12) |
| O1W—Cu1—N1—C1 | −129.43 (9) | N2—C4—C5—N1i | −56.62 (14) |
| N2—Cu1—N1—C5i | −164.78 (9) | O2—C6—C7—C8 | −40.9 (2) |
| N2i—Cu1—N1—C5i | 15.22 (9) | O1—C6—C7—C8 | 137.52 (16) |
| O1W—Cu1—N1—C5i | 104.60 (9) | C7'—C6—C7—C8 | −86.5 (15) |
| N1i—Cu1—N2—C3 | −140.76 (10) | C6—C7—C8—C9 | −66.1 (2) |
| N1—Cu1—N2—C3 | 39.24 (10) | C7—C8—C9—C10 | 174.03 (18) |
| O1W—Cu1—N2—C3 | 129.05 (9) | C8—C9—C10—C11 | −172.59 (19) |
| N1i—Cu1—N2—C4 | −14.63 (9) | C9—C10—C11—C12 | −176.5 (2) |
| N1—Cu1—N2—C4 | 165.37 (9) | O2—C6—C7'—C8' | −105.8 (7) |
| O1W—Cu1—N2—C4 | −104.81 (9) | O1—C6—C7'—C8' | 87.6 (8) |
| C5i—N1—C1—C2 | 179.31 (11) | C7—C6—C7'—C8' | 34.5 (10) |
| Cu1—N1—C1—C2 | 56.13 (14) | C6—C7'—C8'—C9' | 60.9 (11) |
| N1—C1—C2—C3 | −69.50 (16) | C7'—C8'—C9'—C10' | 170.4 (9) |
| C4—N2—C3—C2 | 179.52 (11) | C8'—C9'—C10'—C11' | −175.3 (10) |
| Cu1—N2—C3—C2 | −57.16 (13) | C9'—C10'—C11'—C12' | 174.2 (15) |
| C1—C2—C3—N2 | 70.11 (16) |
Symmetry codes: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1ii | 0.86 (1) | 2.30 (1) | 2.983 (2) | 137 (2) |
| N2—H2···O2 | 0.86 (1) | 2.12 (1) | 2.924 (2) | 156 (2) |
| O1w—H11···O2 | 0.83 (1) | 1.93 (1) | 2.730 (2) | 162 (2) |
| O1w—H12···O2w | 0.83 (1) | 1.95 (1) | 2.777 (2) | 174 (2) |
| O2w—H21···O1iii | 0.83 (1) | 2.02 (1) | 2.833 (2) | 166 (2) |
| O2w—H22···O1ii | 0.84 (1) | 1.91 (1) | 2.743 (2) | 171 (2) |
Symmetry codes: (ii) −x+1, y−1/2, −z+3/2; (iii) x, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5285).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2009). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Hunter, T. M., McNae, I. W., Liang, X., Bella, J., Parsons, S., Walkinshaw, M. D. & Sadler, P. J. (2005). Proc. Natl Acad. Sci. USA, 102, 2288–2292. [DOI] [PMC free article] [PubMed]
- Lindoy, L. F., Mahinay, M. S., Skelton, B. W. & White, A. H. (2003). J. Coord. Chem.56, 1203–1213.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst.43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810025687/bt5285sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810025687/bt5285Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report