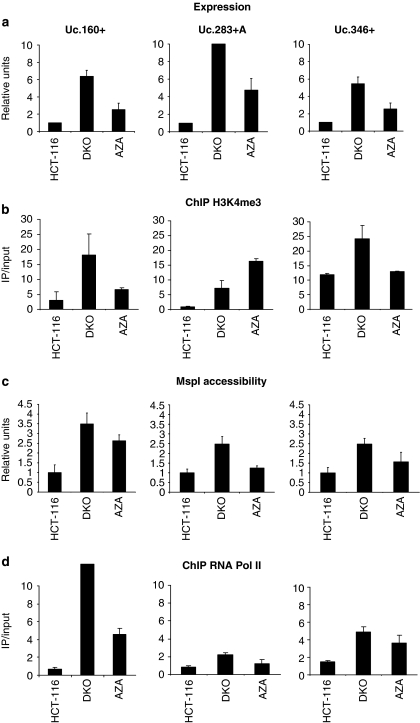
Figure 3.
T-UCR expression and chromatin environment. (a) Expression analyses of T-UCRs by quantitative RT–PCR. Uc.160+, Uc.283+A and Uc.346+ show minimal expression in the hypermethylated HCT-116 cells, while treatment with the DNA-demethylating agent 5-aza-2′-deoxycytidine (AZA) and DKO cells show T-UCR upregulation. (b) Quantitative chromatin immunoprecipitation assay for the histone modification mark trimethylation of lysine 4 of histone H3 (H3K4me3) that is associated with active transcription. The presence of T-UCR CpG island methylation is associated with the lack of H3K4me3 histone, whereas the opposite scenario is observed when DNA demethylation events are present by genetic disruption of the DNMTs (DKO cells) or pharmacological treatment with a DNA-demethylating agent (AZA lane). (c) Chromatin accessibility assay using the MspI restriction enzyme coupled to real-time PCR. The hypermethylated CpG islands of Uc.160+, Uc.283+A and Uc.346+ CpG in HCT-116 are inaccessible to the enzyme, but CpG island demethylation upon 5-aza-2′-deoxycytidine treatment or in DKO cells results in higher accessibility. (d) Quantitative chromatin immunoprecipitation assay for RNA polymerase II (RNA Pol II) shows its absence in T-UCR hypermethylated CpG islands and its enrichment upon DNA hypomethylation events (DKO cells and HCT-116 treated with the DNA-demethylating drug).