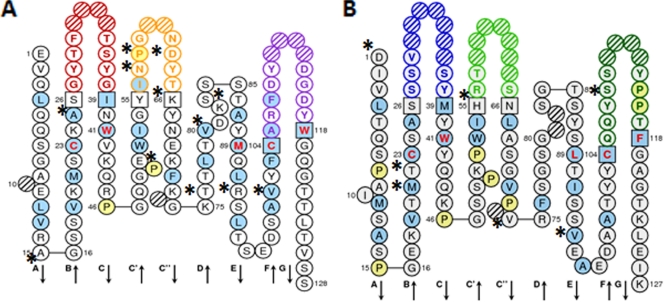
FIG. 6.
IMGT collier de perle graphical 2-dimensional representation of 4PCL2 ScFv. (A) Variable heavy chain (VH). (B) Variable light chain (VL). Collier de perle representations are displayed according to IMGT unique numbering. Asterisks indicate differences between 4PCL2 ScFv and the mouse germ line genes most similar to 4PCL2 ScFv. Hydrophobic amino acids (those with positive hydropathy index values, i.e., I, V, L, F, C, M, and A) and tryptophan (W) are shown in blue circles. All proline (P) residues are shown in yellow circles. The complementarity-determining region (CDR-IMGT) sequences are delimited by amino acids shown in squares (anchor positions), which belong to the neighboring framework region (FR-IMGT). Hachured circles correspond to missing positions according to IMGT unique numbering. Colors of circle outlines indicate regions: in the VH domain, red for CDR1-IMGT, orange for CDR2-IMGT, and purple for CDR3-IMGT; in the VL domain, blue for CDR1-IMGT, green for CDR2-IMGT, and turquoise for CDR3-IMGT. Residues at positions 23 (first cysteine), 41 (tryptophan), 89 (hydrophobic amino acid), 104 (second cysteine), and 118 (hydrophobic amino acid) are conserved and shown in red letters.