Abstract
Noroviruses (NoV) annually cause millions of cases of gastrointestinal disease in the United States. NoV are associated with raw shellfish outbreaks, particularly oysters, which are thought to bioaccumulate NoV particles during the filter-feeding process. NoV outbreaks, however, have also been known to occur from other common-source food-borne vehicles, such as lettuce, frozen raspberries, and salad. In this study, we evaluated romaine lettuce as a potential vehicle for NoV transmission by testing the binding and distribution of NoV to the surface of romaine. Recombinant Norwalk virus-like particles (rNVLP) applied to the surface of romaine lettuce localized as large clusters primarily on the leaf veins. An extract of romaine lettuce leaves in phosphate-buffered saline (PBS) (romaine extract [RE]) bound rNVLP in a dose-dependent manner. RE did not bind rNVLP by histo-blood group antigens (HBGA), nor was RE competitive with rNVLP binding to porcine gastric mucin. These results suggested that non-HBGA molecules in RE bind rNVLP by a binding site(s) that is different from the defined binding pocket on the virion. Extracts of cilantro, iceberg lettuce, spinach, and celery also bound rNVLP. Samples of each of the vegetables spiked with rNVLP and tested with anti-NVLP antibody revealed by confocal microscopy the presence of rNVLP not only on the veins of cilantro but also throughout the surface of iceberg lettuce.
Noroviruses (NoV) cause an estimated 23 million annual cases of sporadic and epidemic gastrointestinal disease in the United States, accounting for one-half to two-thirds of all food-borne illnesses (5, 7). The most common and best characterized route of infection is through the consumption of oysters (4), which are contaminated directly at the source of harvest. Histo-blood group antigens (HBGA), identified as receptors on human gastrointestinal cells to which NoV virions bind (10, 11, 19), are also present on the surfaces of oyster gastrointestinal cells (25). It is postulated, therefore, that during the course of filter feeding, oysters bioaccumulate NoV particles through HBGA binding (15, 25).
Recent outbreaks of food-borne illness have shed light and raised concerns about the growing problem of contamination of foods. This is a concern particularly for cold foods, such as salads, bakery products, and cold cuts, all of which have been associated with NoV outbreaks (21). A recent CDC survey estimated that 58.3% of all food-borne disease outbreaks associated with leafy greens and with confirmed etiologies between 1973 and 2006 were caused by NoV (9).
Contamination of produce can occur at multiple points, including at preharvest by contaminated water sources, contaminated amendments (e.g., raw manure or compost), or wildlife intrusion and at harvest or postharvest by cross-contamination potentially by any of the multiple people who handle the produce (16, 18). Given the low infectious doses of NoV for illness and the long-term persistence of NoV virions (8), it is particularly important to determine whether any of the implicated foods enhance NoV transmission by retaining NoV particles.
We report the results of studies to determine whether leafy greens, specifically romaine lettuce, can serve as a vehicle for NoV transmission through a capability to bind NoV particles. Lettuce is a prime candidate to test because it is well documented that bacteria, such as Salmonella enterica and Escherichia coli O157:H7, are able to bind to, colonize, and contaminate lettuce and other leafy green leaves (13, 14). It is noteworthy that feline calicivirus was reported to transfer between contaminated hands and lettuce or contaminated lettuce and clean hands (1). Furthermore, it was determined that recent outbreaks of food-borne gastroenteritis in Denmark were caused by lettuce contaminated with both enterotoxigenic E. coli and NoV (6). Therefore, it can be hypothesized that lettuce becomes contaminated initially by NoV in the field by some contaminated source, such as irrigation water, or postharvest by cross-contamination by an ill food handler. However, interactions between NoV particles and plant surfaces, specifically those of leafy greens like lettuce, have not been studied extensively. Adherence of NoV to leafy greens by a mechanism that can resist washing could result in the bioaccumulation of NoV from an environment of low concentration (e.g., water) to potentially infectious doses for humans. In the present study, we test the potential for bioaccumulation of NoV on a plant surface by utilizing recombinant Norwalk virus-like particles (rNVLP) to assay and characterize the binding of NoV to romaine lettuce. As a comparison to the results obtained with romaine lettuce, we also assessed the binding of rNVLP to other vegetables.
MATERIALS AND METHODS
Antibodies.
Polyclonal rabbit antiserum against Norwalk virus (R183) was kindly provided by P. Reilly (Wyeth Research, Pearl River, NY); R183 activity has been described previously (24, 25). Goat anti-rabbit immunoglobulin conjugated to Alexa 488 and goat anti-rabbit immunoglobulin conjugated to alkaline phosphatase (AP; Invitrogen, Carlsbad, CA) were used to detect R183. Monoclonal antibodies were used to detect various HBGA; antibodies designated BG1 to BG8 are specific for a common blood group antigen precursor glycoprotein (BG1) and type A, type B, type H1, type Lewis a, type Lewis b, type Lewis x, and type Lewis y HBGA, respectively (Signet Labs, Dedham, MA). BG antibodies were detected with goat anti-mouse immunoglobulin conjugated to alkaline phosphatase (Zymed Labs, South San Francisco, CA).
Cells and virus preparation.
Sf9 cells were grown in a suspension culture at 28°C in TNM-FH insect media (Gemini Bio-Products, West Sacramento, CA) supplemented with 10% fetal bovine serum (JR Scientific, Woodland, CA) and 0.1% Pluronic F-68, a cell stabilizer and surfactant (Sigma-Aldrich, St. Louis, MO). For infection, cells were transferred to ESF-921 protein-free insect cell culture media (Expression Systems, Davis, CA). Recombinant Norwalk virus-like particles (rNVLP) were generated by expressing the Norwalk virus capsid (NCBI reference NC_001959.2) from the BestBac recombinant baculovirus expression system (Expression Systems). At 5 to 6 days postinfection, the infected cell suspension was centrifuged for 10 min at 5,000 rpm (approximately 3,300 × g) in an Eppendorf 5810 R centrifuge. The supernatant was transferred to a fresh container, stored at 4°C, and used for the enzyme-linked immunosorbent assay (ELISA). For immunofluorescence microscopy, rNVLP were further purified by ultracentrifugation through a 30% sucrose cushion in a Beckman-Coulter ultracentrifuge at 25,000 rpm in a SW28 rotor (82,500 × g) at 15°C for 2 h. The resulting pellet was resuspended in phosphate-buffered saline (PBS; 100 μM, pH 7.5) and stored at 4°C.
Extract of romaine lettuce leaves.
Heads of romaine lettuce were purchased from a local grocery store and stored in the lab at 4°C. For microscopy, strips were cut from the ends of leaves, flattened onto precleaned glass microscope slides, and then taped down at the edges, exposing approximately 1 to 2 cm2 of romaine. For ELISA, crushed romaine extract (RE) was prepared by cutting the top half of a leaf, weighing it, and placing it into a 50-ml conical vial. An equal amount of PBS (wt/vol) was added to the conical vial, and the blunt end of a cell spreader was used to crush the romaine leaf in the tube for 2 min. The vial was then vortexed for 1 min, and the liquid phase was transferred to a fresh conical vial and centrifuged at 10,000 × g for 10 min. The supernatant was transferred to a fresh tube and stored at 4°C for up to 2 days.
The other vegetables tested were prepared similarly but with modifications noted below. After green onions were crushed, it was necessary to cool the liquid phase at 4°C for 30 min prior to centrifugation. For raspberries, an initial round of centrifugation was performed after vortexing and prior to transferring the liquid phase to a fresh tube. Celery stalks were crushed using a sterile mortar and pestle, and the juice was transferred to a conical vial with PBS equal to the weight of the initial stalk.
All vegetable preparations involved samples taken from multiple parts of the same retail sample as well as different vegetable samples purchased over a period of several months. ELISA and microscopy experiments were performed at least in triplicate with samples taken from vegetables purchased at different times to represent different field conditions.
Fluorescence microscopy.
Strips of romaine on glass slides were blocked with 10% skim milk in PBS at 4°C overnight and then washed for 5 min in PBS. Partially purified rNVLP (100 μg total protein in 100 μl PBS) or PBS was applied to the surface of the romaine, which was then topped by a coverslip to evenly distribute the virus on the surface of the lettuce. After 1 h of incubation, the slides were washed in PBS plus 1% Tween 80 (PBS-T) for 5 min. Anti-Norwalk virus antibody (R183; 1:2,000 in PBS) was applied to the surface, covered, and incubated at room temperature for 1 h, followed by a 5-min wash in PBS-T. Alexa 488-conjugated secondary antibody (1:100 in PBS) was applied to the surface, covered, and incubated at room temperature for 1 h, followed by a 5-min wash in PBS-T and a 5-min wash in PBS. Thirty microliters of PBS was applied to the surface of the romaine, covered by a coverslip, and viewed on a Leica dissecting microscope equipped for fluorescence microscopy or a Leica confocal laser scanning microscope.
Iceberg lettuce, cilantro, and spinach strips on glass slides were prepared as described above for microscopy. Celery stalks were assayed along their concave surface, and all solutions were spread across the surface using a pipette tip to evenly distribute the solution.
Measurement of the binding of rNVLP to RE by ELISA.
To test the binding of rNVLP, Nunc Immuno modules were incubated at 4°C overnight with a coating of 100 μl of PBS, porcine gastric mucin (PGM) (100 μg/ml unless otherwise specified; Sigma-Aldrich), or vegetable extract (RE was diluted 1:5 in PBS, unless otherwise specified; all other extracts were tested undiluted). To determine whether proteins were involved in rNVLP binding, PGM and RE were boiled for 5 min prior to being applied to wells. Unbound extract material was removed by decanting, and wells were blocked with 10% skim milk in PBS at 37°C for 2 h. Plates were washed 2 times with Tris-buffered saline (TBS) plus 1% Tween 20 (TBS-T) using a BioTek plate washer, and 100 μl PBS or 100 μl rNVLP (1 μg/μl of total protein) was added to wells and incubated at 37°C for 1 h. To determine whether carbohydrates were involved in rNVLP binding, 100 μl of potassium periodate (4 mg/ml; Sigma-Aldrich) was added to wells, and plates were incubated for 30 min at 37°C prior to the addition of rNVLP. For competition assays, PBS or rNVLP were preincubated with competitor at 37°C for 30 min immediately prior to being added to wells. After 1 h of incubation with rNVLP, plates were washed 2 times with TBS-T, 100 μl R183 (1:2,500 in PBS) was added to wells, and plates were incubated at 37°C for 1 h. Plates were washed 2 times with TBS-T, 100 μl of AP-conjugated secondary antibody (1:3,000 in PBS) was added to each well, and plates were incubated at 37°C for 1 h. Plates were washed 3 times with TBS-T, and 100 μl p-nitrophenyl phosphate (PNPP) disodium salt (1 mg/ml in diethanolamine substrate buffer; Pierce) was added to each well. Plates were allowed to develop for up to 100 min, and absorbance readings at 405 nm were recorded every 20 min on a SpectraMax ELISA reader.
To test romaine for the presence of HBGA-like motifs, Nunc Immuno modules were incubated at 4°C overnight with 100 μl of the extract coating solution, decanted, and blocked with skim milk as described above. Plates were washed 2 times with TBS-T, 100 μl of individual BG antibodies (1:50 in PBS) were added to each well, and plates were incubated at 37°C for 1 h. Plates were then washed 2 times with TBS-T, 100 μl of AP-conjugated immunoglobulin was added to each well, and plates were incubated at 37°C for 1 h. Plates were washed 3 times in TBS-T, and the antibody binding was determined by addition of PNPP, as described above.
Statistics.
For microscopy, binding was tested using strips from at least three different vegetable samples bought at different times of the year. For the ELISA, each plate contained at least 3 wells for each treatment, and each treatment was repeated at least three times with vegetable samples bought at different times. Each plate also contained the following two internal negative controls: rNVLP in PBS-coated wells and wells coated with vegetable extract without added rNVLP. The negative control with the higher optical density (OD) value was used as the nonspecific binding value, equal to 1. Competitive binding values (see Fig. 3) were compared by one-way analysis of variance (ANOVA), using the program SigmaStat.
RESULTS
Localization of rNVLP binding to romaine lettuce.
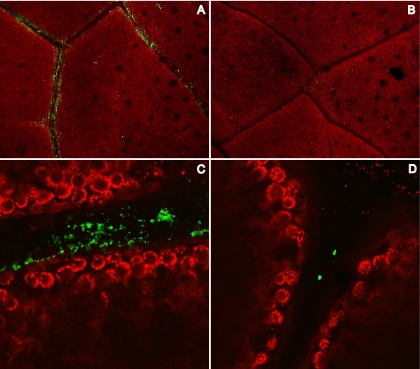
To determine if rNVLP binding could be observed directly on romaine lettuce, rNVLP was applied to lettuce and stained with R183 anti-Norwalk virus antibody followed by an Alexa 488-conjugated secondary antibody. Using the natural fluorescence/autofluorescence of romaine for contrast, rNVLP were visualized under a fluorescent dissecting microscope and a confocal microscope. rNVLP clusters remained on romaine leaves after being washed and were localized primarily to the veins of the romaine leaf, suggesting possible specificity of rNVLP binding (Fig. 1A and C). Romaine samples without rNVLP added had minimal background antibody staining (Fig. 1B and D). Furthermore, romaine leaves without exposure to antibody showed no staining, regardless of the presence of rNVLP (data not shown). Occasional antibody staining was observed on the samples without rNVLP added (Fig. 1D), possibly due to cross-reactivity of one of the antibodies with an unknown leaf antigen, nonleaf material, or an unknown organism bound to the leaf. Nevertheless, significant differences were observed in the overall amount of rNVLP detected compared to that detected with the no-rNVLP control.
FIG. 1.
Strips of romaine lettuce were viewed under a dissecting microscope (A and B) or a confocal microscope (C and D), after the application of rNVLP (A and C) or PBS (B and D), followed by antibody staining. Red, autofluorescence of romaine leaves; green, rNVLP.
Dose-dependent binding of rNVLP to romaine lettuce extract.
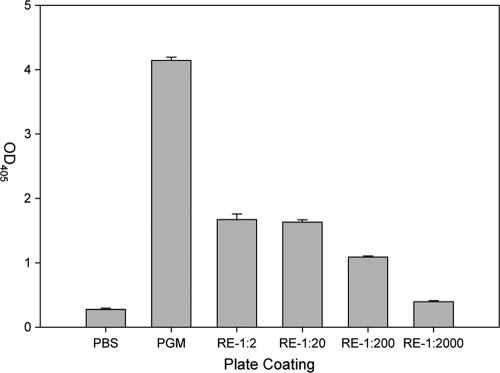
The specific localization of rNVLP binding to the surface of romaine suggested that romaine might have a ligand on the surface that serves as a receptor for rNVLP. Wells coated with a 50% dilution of romaine extract (RE) bound a 6-fold-larger amount of rNVLP (OD at 405 nm [OD405] value of 1.670 ± 0.085) compared to that bound by uncoated wells (OD405 value of 0.277 ± 0.019) (Fig. 2). In contrast, wells coated with the PGM positive control bound a 15-fold-larger amount of rNVLP (OD405 value of 4.139 ± 0.054) compared to that bound by uncoated wells. rNVLP binding appeared to be dose dependent over a 3-log range of diluted extract, although binding at a 1:2,000 dilution of RE had a positive-to-negative (P/N) ratio of less than 2 (OD405 of 0.395 ± 0.018).
FIG. 2.
Wells of a plate were coated with PBS, PGM, or the listed dilution amount of RE, and rNVLP binding was assayed by ELISA. Error bars represent standard deviations (SD).
Characterization of molecules involved in rNVLP binding to RE.
RE was compared to PGM in terms of binding of rNVLP to determine whether the binding domains on rNVLP were similar for the two types of samples. PGM was used as a positive control, since HBGA has been characterized as the receptor for human NoV and receptor binding has been localized to the HBGA-binding pocket of the protruding domain of NoV particles. By ELISA, antibodies against precursor-type, ABO-type, and Lewis-type HBGA did not bind to RE-coated wells, indicating no HBGA-like sugars of these types present in RE (data not shown).
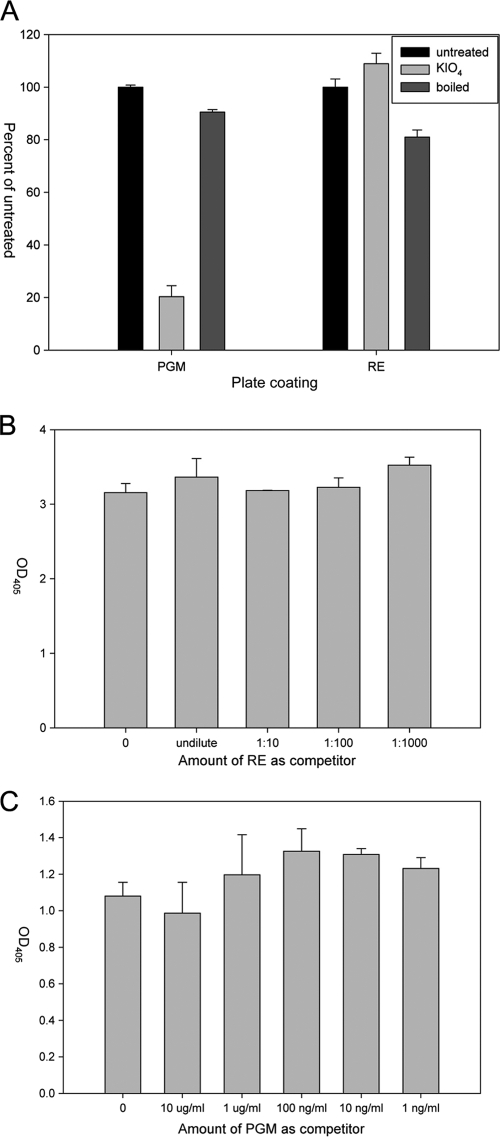
The RE tested did not have any cross-reacting HBGA, but we wanted to determine whether rNVLP could bind to other protein or sugar moieties contained on or within romaine lettuce. Therefore, the RE was treated by being boiled or with potassium periodate (KIO4) to denature proteins or oxidize carbohydrates, respectively. PGM was used as a positive binding control. rNVLP binding to PGM was affected by treatment with KIO4 but not by being boiled, confirming carbohydrate involvement in rNVLP binding to PGM (Fig. 3A). Binding of rNVLP to RE, however, was affected by both KIO4 and by boiling RE by a small, but statistically significant, amount. KIO4 treatment resulted in a minor increase in binding (8.9 ± 4.0%) relative to untreated RE (P < 0.05). Boiling the RE resulted in a decrease of rNVLP binding (19.3 ± 5.0%) relative to untreated RE (P < 0.01), with a range of 14 to 25% (Fig. 3A).
FIG. 3.
Comparison between rNVLP binding to PGM and RE was assayed by ELISA. (A) The involvement of sugar and protein moieties in rNVLP binding was determined by treatment of PGM or RE with KIO4 or after being boiled. (B) Competition assay in which rNVLP were preincubated with the listed dilution amount of RE prior to incubation on PGM-coated plates. (C) rNVLP were preincubated with the listed amount of PGM prior to incubation on RE-coated plates. Error bars represent SD.
Competition assays were also performed to determine if RE and PGM interacted with the same binding site of rNVLP. No inhibition was observed with rNVLP preincubated with up to 10 μg/ml PGM before addition to RE-coated wells or with rNVLP preincubated with various dilutions of RE before addition to PGM-coated wells (P > 0.05) (Fig. 3B and C).
Binding of rNVLP to other vegetable extracts.
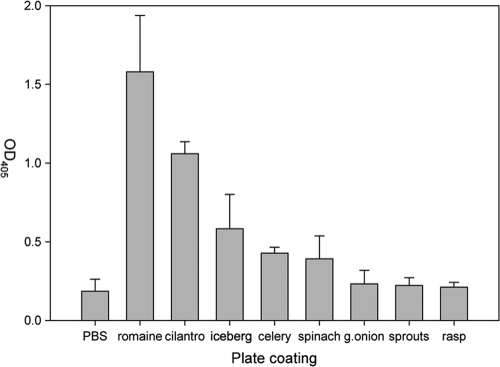
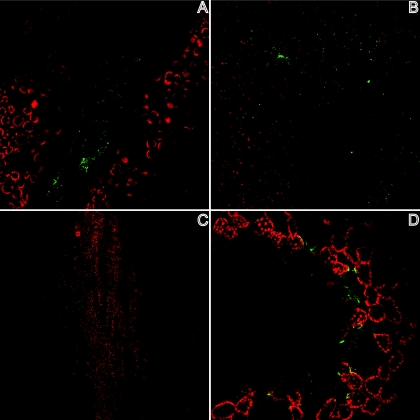
The results of experiments performed on romaine lettuce stimulated us to survey other vegetable extracts for binding of rNVLP. Extracts prepared from cilantro, iceberg lettuce, celery, spinach, green onions, clover sprouts, and raspberries were used to coat ELISA wells to measure rNVLP binding (Fig. 4). Using a P/N ratio of 2 as the positive binding cutoff, we determined that cilantro (OD405 of 1.060 ± 0.075), iceberg lettuce (OD405 of 0.583 ± 0.218), celery (OD405 of 0.427 ± 0.038), and spinach (OD405 of 0.391 ± 0.146) bound rNVLP at significant levels compared to that of PBS (OD405 of 0.186 ± 0.076). However, none of these vegetable extracts bound rNVLP as well as RE (OD405 of 1.580 ± 0.358). Microscopy revealed that rNVLP localized to the veins of cilantro, similar to the localization observed for romaine lettuce. In contrast, iceberg lettuce bound rNVLP over the whole leaf, whereas no rNVLP on the surface of celery was observed. Finally, rNVLP appeared to bind to spinach at specific locations that appeared to be depressions on the leaf (Fig. 5).
FIG. 4.
Wells of a plate were coated with extracts from each of the different listed vegetables, and rNVLP binding was assayed by ELISA. Error bars represent SD.
FIG. 5.
rNVLP binding to the surfaces of cilantro (A), iceberg lettuce (B), celery (C), and spinach (D) was assayed by confocal microscopy. Red, autofluorescence of vegetable; green, rNVLP.
DISCUSSION
The results of this study indicate that rNVLP can bind to romaine lettuce. Even rNVLP applied evenly throughout the surfaces of romaine leaves appeared by confocal microscopy to localize in clusters on veins rather than with an even distribution throughout the romaine leaf. The localization of the binding of rNVLP along romaine leaf veins implies that binding may involve a specific ligand(s) on the leaf surface.
Veins of leaves have a thinner waxy coat than the overall leaf surface and bind naturally occurring epiphytic bacteria along veins in a manner similar to that of rNVLP shown in this study (2). Several species of bacteria are able to express HBGA-like sugars on their lipopolysaccharide O-chains (20, 27); therefore, it is possible that rNVLP might have bound to lipopolysaccharide (LPS) or some other component of natural bacteria bound to veins (2). Our preliminary results suggest this is not the case, but we cannot yet eliminate this as a possible mechanism of binding to components in RE or on the leaf surface. For example, a surface wash of a romaine leaf was plated on Luria-Bertani agar plates, and the three most common bacteria (based on colony morphology) were isolated. None of these bacteria used to coat wells of an ELISA plate could bind rNVLP (data not shown). Furthermore, the RE preparation included a 10,000 × g centrifugation step that should eliminate most, if not all, of the bacterial cells in the extract, minimizing them as possible receptors for rNVLP. Finally, rNVLP binding to RE did not mimic rNVLP binding to HBGA, and anti-HBGA antibodies did not bind to RE, implying that bacteria with HBGA-like sugars on the surface were not present in RE to facilitate rNVLP binding.
Since rNVLP do not appear to bind bacteria on the romaine surface, it seemed possible that rNVLP were binding to plant molecules exposed at the veins. It is difficult to test the vein region only, so the binding of rNVLP to romaine lettuce was further assessed with RE. Binding of rNVLP to RE molecules with and without inhibitors was assessed by an ELISA developed previously to test NoV binding to various substrates (25). Binding was dose dependent over a 1,000-fold range to RE and was relatively specific (6-fold higher than that of the negative control). rNVLP binding to RE was reproducible with multiple preparations from different leaves and different heads of retail romaine lettuce. However, the extent of binding relative to the negative control was variable between preparations, ranging from P/N ratios of 3.5 to 10. The variability in binding between preparations could be the result of differing amounts of a binding factor released during extraction or of differences in the physical or anatomical factors in the different heads of lettuce. Additional research will be needed to determine the exact nature of the receptors binding rNVLP in this study.
HBGA are recognized receptors for human NoV, and binding has been mapped to the outer pocket of the P domain protrusions on the Norwalk virus shell (3, 22). RE did not appear to contain any HBGA-like sugars based on the lack of binding of specific anti-HBGA antibodies by ELISA. Currently, it is unclear if ligands other than HBGA are involved in NoV binding. The binding of rNVLP to romaine was therefore compared to the binding of rNVLP to PGM, which contains type A, type O, and type Lewis b HBGA and to which Norwalk virus is known to bind (24). Unlike PGM, which binds to rNVLP primarily via sugar moieties, as reflected by abolished binding activity after oxidation of PGM (24), RE binding of rNVLP was increased slightly by oxidation but decreased slightly but significantly by being boiled. This suggests that proteins might have some role in rNVLP binding to RE, whereas sugars do not play a role and may serve to mildly inhibit binding. However, the leaf phylloplane is composed of epidermal cells covered by cuticle, which is a mixture of cutin, polysaccharides, and a variety of waxes composed of long-chain fatty acids (17). For example, a wax (stigmasterol) extracted from cilantro leaves was shown to bind Salmonella enterica cells (see Fig. 2.3F in reference 17). Whether a single protein or a more complex set of glycoconjugates, proteins, waxes, and/or other molecules provide multiple binding receptors on romaine remains unclear. The results of these experiments, however, do imply that some molecules in romaine lettuce, unlike HBGA, might be involved in the binding of rNVLP, molecules which have not been described previously for human NoV.
Competition assays were performed between RE and PGM to determine if ligands in RE might bind to the outer pocket of the P domain of rNVLP, shown previously to bind to HBGA receptors. A dilution series of RE preincubated with rNVLP had no effect on rNVLP binding to PGM. Similarly, when PGM was preincubated with rNVLP prior to rNVLP incubation with RE, PGM at concentrations of up to 10 μg/ml did not compete against rNVLP binding to RE. The lack of competition between PGM and RE for rNVLP binding implies that RE does not utilize the HBGA binding pocket of rNVLP, which also has not been characterized previously for the attachment of Norwalk virus.
Based on the results of rNVLP binding to romaine lettuce, binding of rNVLP to a variety of vegetables was assayed. rNVLP were determined to bind to extracts from cilantro, iceberg lettuce, celery, and spinach but not to green onions, clover sprouts, and raspberries. This binding was weaker than binding to RE, and when further analyzed by microscopy, only cilantro and iceberg lettuce were observed to consistently bind rNVLP. rNVLP localized to the veins of cilantro, in a pattern that mimicked the localization of rNVLP on romaine lettuce, suggesting that cilantro may also contain a specific ligand that is able to bind rNVLP and that this ligand may be present along the veins where the waxy cuticle is thinner or absent. Iceberg lettuce, on the other hand, was determined to bind rNVLP along the entire surface. This suggests that a ligand is present along the length of the iceberg leaf surface and/or that binding is less specific than to other vegetable locations. In contrast, celery stalks did not appear to bind rNVLP along the surface, possibly due to diffuse binding on the surface or a lack of clustering of particles, which could make the signal too weak to detect. Another possibility is that the binding observed by ELISA is due to the release of a factor when preparing the extract that is internal to the celery stalk normally and is not present on the stalk surface to allow rNVLP binding. Finally, rNVLP bound weakly along the surface of a spinach leaf, or perhaps, rNVLP were trapped in pockets along the surface.
During the course of preparing the manuscript, a similar study was performed looking at murine NoV (MNoV) binding to romaine lettuce. Wei and colleagues (26) determined that MNoV binds to the surface, stomata, and open cuts on romaine, though they could not confirm that binding was statistically significant. Furthermore, they demonstrated that various biosolids increased the binding of MNoV to romaine. As norovirus contamination is likely caused by human fecal contamination, this result is relevant to theories about preharvest contamination of produce, including adjacent contaminated watersheds or biosolid soil amendments. The major difference between the cited study and our study is the use of MNoV rather than recombinant human norovirus. The binding characteristics of human NoV to HBGA have been characterized with live virions of several species as well as virus-like particles (3, 22), whereas MNoV appears to use ganglioside-linked terminal sialic acids as receptors on macrophages (23). Furthermore, the crystal structure of the MNoV capsid demonstrates a structure very different from those of human NoV and other caliciviruses (12), which implies that the binding properties of MNoV also could be different than those of other human NoV and caliciviruses. Since MNoV can be amplified in cell culture, Wei and colleagues used SYBR green-tagged quantifiable amounts of viral particles, facilitating visualization of single virions bound to the romaine leaf (26). In contrast, our study utilized human rNVLP to look for general binding patterns on romaine and determined that rNVLP bound preferentially to the veins of romaine leaves. The differences in the binding and pathogenicity of MNoV compared to those of human NoV, however, emphasize that human NoV is an appropriate choice for studies related to improved detection, tracking, and biology of NoV in food production environments and for information relevant to NoV illness.
In conclusion, we have shown that human rNVLP bind to the surfaces of romaine lettuce. The localization pattern of rNVLP on romaine suggests that specific ligands may be available on the leaf surface to bind rNVLP. We hypothesize that ligands enhance the bioaccumulation of NoV particles on romaine, similar to processes that occur in oysters (15, 25). Bioaccumulation may also occur on other leafy greens, including iceberg lettuce and cilantro. Fundamental research will be needed to identify and characterize the potential binding ligand(s) and to determine whether other leafy green types and their cultivars can bind rNVLP and intact NoV. Additionally, determining how the virions of different NoV species bind to produce and the effect of biosolids and other cocontaminants on binding will be important for elucidating the health risks to produce in public. The numerous outbreaks and illnesses associated with NoV-contaminated produce emphasize the need to better understand NoV biology related to fresh produce in general and leafy greens in particular. The most important goal, however, is to determine whether NoV is present in produce production environments and whether preharvest contamination with NoV could be contributing to produce associated outbreaks.
Acknowledgments
We acknowledge David Yang for excellent technical support and Anna Bates for assistance with confocal microscopy.
This work was supported by the USDA, Agricultural Research Service, CRIS project 5325-42000-044.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Bidawid, S., N. Malik, O. Adegbunrin, S. A. Sattar, and J. M. Farber. 2004. Norovirus cross-contamination during food handling and interruption of virus transfer by hand antisepsis: experiments with feline calicivirus as a surrogate. J. Food Prot. 67:103-109. [DOI] [PubMed] [Google Scholar]
- 2.Brandl, M. T., and R. E. Mandrell. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bu, W., A. Mamedova, M. Tan, M. Xia, X. Jiang, and R. S. Hegde. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, P. K., D. K. Wong, T. W. Chung, and W. W. Lim. 2005. Norovirus contamination found in oysters worldwide. J. Med. Virol. 76:593-597. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, N. A., D. A. Bergmire-Sweat, K. J. Schwab, K. A. Hendricks, S. Reddy, S. M. Rowe, R. L. Fankhauser, S. S. Monroe, R. L. Atmar, R. I. Glass, and P. Mead. 2000. A foodborne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 181:1467-1470. [DOI] [PubMed] [Google Scholar]
- 6.Ethelberg, S., M. Lisby, B. Bottiger, A. C. Schultz, A. Villif, T. Jensen, K. E. Olsen, F. Scheutz, C. Kjelso, and L. Muller. 2010. Outbreaks of gastroenteritis linked to lettuce, Denmark, January 2010. Eurosurveillance 15:1-3. [PubMed] [Google Scholar]
- 7.Glass, R. I., J. Bresee, B. Jiang, J. Gentsch, T. Ando, R. L. Fankhauser, J. Noel, U. D. Parashar, B. Rosen, and S. S. Monroe. 2001. Gastroenteritis viruses: an overview. Novartis Found. Symp. 238:5-19. [DOI] [PubMed] [Google Scholar]
- 8.Glass, R. I., U. D. Parashar, and M. K. Estes. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman, K. M., T. L. Ayers, and M. Lynch. 2008. Foodborne disease outbreaks associated with leafy greens, 1973-2006, p. 27. Abstr. Int. Conf. Emerg. Infect. Dis., Atlanta, GA, 16 to 19 March 2008.
- 10.Huang, P., T. Farkas, S. Marionneau, W. Zhong, N. Ruvoen-Clouet, A. L. Morrow, M. Altaye, L. K. Pickering, D. S. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19-31. [DOI] [PubMed] [Google Scholar]
- 11.Hutson, A. M., R. L. Atmar, D. M. Marcus, and M. K. Estes. 2003. Norwalk virus-like particle hemagglutination by binding to histo-blood group antigens. J. Virol. 77:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katpally, U., C. E. Wobus, K. Dryden, H. W. Virgin IV, and T. J. Smith. 2008. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J. Virol. 82:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, J. K., and M. A. Harrison. 2008. Transfer of Escherichia coli O157:H7 to romaine lettuce due to contact water from melting ice. J. Food Prot. 71:252-256. [DOI] [PubMed] [Google Scholar]
- 14.Kroupitski, Y., R. Pinto, M. T. Brandl, E. Belasouv, and S. Sela. 2009. Interaction of Salmonella enterica with lettuce leaves. J. Appl. Microbiol. 106:1876-1885. [DOI] [PubMed] [Google Scholar]
- 15.Le Guyader, F., F. Loisy, R. L. Atmar, A. M. Hutson, M. K. Estes, N. Ruvoen-Clouet, M. Pommepuy, and J. Le Pendu. 2006. Norwalk virus-specific binding to oyster digestive tissues. Emerg. Infect. Dis. 12:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch, M. F., R. V. Tauxe, and C. W. Hedberg. 2009. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 137:307-315. [DOI] [PubMed] [Google Scholar]
- 17.Mandrell, R., L. Gorski, and M. Brandl. 2006. Attachment of microorganisms to fresh produce, p. 34-61. In G. M. Sapers, J. R. Gorny, and A. E. Yousef (ed.), Microbiology of fruits and vegetables. CRC, Taylor and Francis, Boca Raton, FL.
- 18.Mara, D., and A. Sleigh. 2010. Estimation of norovirus infection risks to consumers of wastewater-irrigated food crops eaten raw. J. Water Health 8:39-43. [DOI] [PubMed] [Google Scholar]
- 19.Marionneau, S., N. Ruvoen, B. Le Moullac-Vaidye, M. Clement, A. Cailleau-Thomas, G. Ruiz-Palacois, P. Huang, X. Jiang, and J. Le Pendu. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasko, D. A., G. Wang, M. A. Monteiro, M. M. Palcic, and D. E. Taylor. 2000. Synthesis of mono- and di-fucosylated type I Lewis blood group antigens by Helicobacter pylori. Eur. J. Biochem. 267:6059-6066. [DOI] [PubMed] [Google Scholar]
- 21.Seymour, I. J., and H. Appleton. 2001. Foodborne viruses and fresh produce. J. Appl. Microbiol. 91:759-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taube, S., J. W. Perry, K. Yetming, S. P. Patel, H. Auble, L. Shu, H. F. Nawar, C. H. Lee, T. D. Connell, J. A. Shayman, and C. E. Wobus. 2009. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 83:4092-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian, P., A. H. Bates, H. M. Jensen, and R. E. Mandrell. 2006. Norovirus binds to blood group A-like antigens in oyster gastrointestinal cells. Lett. Appl. Microbiol. 43:645-651. [DOI] [PubMed] [Google Scholar]
- 25.Tian, P., A. L. Engelbrektson, X. Jiang, W. Zhong, and R. E. Mandrell. 2007. Norovirus recognizes histo-blood group antigens on gastrointestinal cells of clams, mussels, and oysters: a possible mechanism of bioaccumulation. J. Food Prot. 70:2140-2147. [DOI] [PubMed] [Google Scholar]
- 26.Wei, J., Y. Jin, T. Sims, and K. E. Kniel. 2010. Manure- and biosolids-resident murine norovirus 1 attachment to and internalization by Romaine lettuce. Appl. Environ. Microbiol. 76:578-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi, W., J. Shao, L. Zhu, M. Li, M. Singh, Y. Lu, S. Lin, H. Li, K. Ryu, J. Shen, H. Guo, Q. Yao, C. A. Bush, and P. G. Wang. 2005. Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J. Am. Chem. Soc. 127:2040-2041. [DOI] [PubMed] [Google Scholar]