Abstract
Brucella is a Gram-negative bacterium that causes a worldwide-distributed zoonosis. The genus includes smooth (S) and rough (R) species that differ in the presence or absence, respectively, of the O-polysaccharide of lipopolysaccharide. In S brucellae, the O-polysaccharide is a critical diagnostic antigen and a virulence determinant. However, S brucellae spontaneously dissociate into R forms, a problem in antigen and S vaccine production. Spontaneous R mutants of Brucella abortus, Brucella melitensis, and Brucella suis carried the chromosomal scar corresponding to genomic island 2 (GI-2) excision, an event causing the loss of the wboA and wboB O-polysaccharide genes, and the predicted excised circular intermediate was identified in B. abortus, B. melitensis, and B. suis cultures. Moreover, disruption of a putative phage integrase gene in B. abortus GI-2 caused a reduction in O-polysaccharide loss rates under conditions promoting S-R dissociation. However, spontaneous R mutants not carrying the GI-2 scar were also detected. These results demonstrate that the phage integrase-related GI-2 excision is a cause of S-R brucella dissociation and that other undescribed mechanisms must also be involved. In the R Brucella species, previous works have shown that Brucella ovis but not Brucella canis lacks GI-2, and a chromosomal scar identical to those in R mutants was observed. These results suggest that the phage integrase-promoted GI-2 excision played a role in B. ovis speciation and are consistent with other evidence, suggesting that this species and B. canis have emerged as two independent lineages.
Brucellosis is one of the most important bacterial zoonotic diseases. This infection is caused by the members of the genus Brucella, a group of Gram-negative, intracellular facultative microorganisms (13). Brucella abortus, Brucella melitensis, Brucella suis, Brucella neotomae, Brucella ceti, and Brucella pinnipedialis are denominated smooth (S) brucellae because their colonies have glossy, bluish surfaces that contrast with the rough (R), granular aspect of the Brucella canis and Brucella ovis colonies. Moreover, S brucellae had a marked tendency to yield mixtures of S and R colonies, a phenomenon known as Brucella S-R dissociation (4). These colony phenotypes respectively relate to the presence or absence of the O-polysaccharide of lipopolysaccharide (LPS) (1, 14), and since this structure is a major diagnostic antigen and virulence factor, S-R dissociation has drawn considerable attention (1, 4, 14, 21). Indeed, S-R dissociation is a problem in the production of diagnostic antigens and vaccines (3). Although first observed over 70 years ago, the genetic bases of Brucella S-R dissociation have not been elucidated as of yet.
Up to now, 10 Brucella O-polysaccharide genes (putatively coding for enzymes furnishing sugar precursors, glycosyltransferases, and the O-chain transport system) have been identified, and the corresponding mutants show the R phenotype (1, 9). These genes are located in two major genetic regions, wbo and wbk. The wboA and wboB genes (encoding O-polysaccharide glycosyltransferases) are part of a genomic island (GI) known as GI-2 (17, 18). This island contains a putative phage integrase gene that could promote recombination between flanking repeat sequences, thus causing a deletion that would include those two glycosyltransferases. Accordingly, GI-2 could be unstable, and this could account at least in part for the tendency of S brucellae to dissociate into R variants. Here, we report the characterization of spontaneous R Brucella mutants carrying the complete deletion of GI-2 and the detection of the corresponding excised circular intermediate. We also provide experimental evidence for the role of the GI-2 phage integrase gene in promoting S-R dissociation.
MATERIALS AND METHODS
Bacterial strains, growth conditions, phenotype identification, and plasmids.
The strains used in this study are listed in Table 1. To prepare the inoculum for dissociation experiments, bacteria were grown in Brucella agar plates (Becton Dickinson, Sparks, MD) for 48 h at 37°C, and a loopful was transferred to a 50-ml Erlenmeyer flask with 10 ml of Brucella broth (Becton Dickinson). The broth was incubated with shaking until bacteria reached stationary phase (48 h at 37°C), an aliquot was adjusted at an A750 of 0.109 ± 0.005 using sterile broth, and 100 μl was used to inoculate a new 50-ml Erlenmeyer flask with 10 ml of broth supplemented with McIlvaine's buffer (pH 6.6) (4). This broth was incubated statically for 10 days at 37°C, bacterial colony counts were obtained by plating serial 10-fold dilutions, and S-R dissociation was assessed by using the crystal violet dye exclusion test (27). For each strain, two plates with approximately 1,500 colonies per plate (the limit for obtaining isolated colonies in 36 to 48 h; longer times produced confluent growth) were examined, and three independent experiments were performed. For cloning, Escherichia coli was grown in Luria-Bertani broth (Becton Dickinson) supplemented with kanamycin (50 μg/ml) or chloramphenicol (20 μg/ml). The plasmids used in this work are listed in Table S1 in the supplemental material.
TABLE 1.
Bacterial strains
| Strain | Characteristics | Source or referencea |
|---|---|---|
| B. melitensis 16M | B. melitensis biovar 1 reference strain; S | SAG |
| B. suis 1330 | B. suis biovar 1 reference strain; S | SAG |
| B. abortus 2308 | B. abortus biovar 1 reference strain; S | SAG |
| B. abortus 2308 Nalr | Nalidixic acid-resistant B. abortus 2308; S | UNAV |
| B. ovis 63/290 | Reference strain; R | SAG |
| B. abortus B10 | B. abortus biovar 1, Chilean field isolate; S | 11 |
| B. abortus R1 | Spontaneous ΔGI-2 mutant derived from strain 2308; R | This study |
| B. abortus R6 | Spontaneous ΔGI-2 mutant derived from strain B10; R | This study |
| B. abortus int::Kmr | int mutant (BAB1_0983), 2308 Nalr derivative; S | This study |
| B. abortus int::Kmr/int | int::Kmr mutant complemented with plasmid pMR10int (carrying a copy of int); S | This study |
| E. coli TOP10F | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli OmniMAX 2-t1 | F′ [proAB+lacIq lacZΔM15 Tn10(TetR) Δ(ccdAB)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80(lacZ)ΔM15 Δ(lacZYA-argF) U169 endA1 recA1 supE44 thi-1 gyrA96 relA1 tonA panD | Invitrogen |
| E. coli S17-1λpir | Mating strain with plasmid RP4 inserted into the chromosome | 20 |
SAG, Servicio Agrícola y Ganadero, Chile; UNAV, Universidad de Navarra, Spain.
Genetic analysis of spontaneous R mutants.
Identification of GI-2 scars in spontaneous R mutants was carried out first by PCR analysis (see below) of individual crystal violet-positive colonies (approximately 80 R colonies for B. abortus 2308 of a total of 3,000 colonies). The frequency of GI-2 deletion was then calculated with respect to the total number of R colonies examined. In addition, the numbers of GI-2 scars in the inocula used for plating were examined by quantitative PCR (qPCR) (see below). In this case, the frequency of detection of the GI-2 deletion was calculated with respect to the number of copies of the genome.
Sequence and restriction analyses.
The complete sequences of B. abortus 2308 and B. ovis ATCC 25840 chromosome I were downloaded from GenBank (accession numbers AM040264 and CP000708, respectively). Similarity searches were performed by BLAST (http://www.ncbi.nlm.nih.gov/BLAST). In silico restriction analysis of GI-2 was performed with NEBcutter v2.0 (http://tools.neb.com/NEBcutter2/index.php). Primers (see Table S2 in the supplemental material) were generated with Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi). DNA sequencing by the dideoxy method was performed at Macrogen Inc., Seoul, South Korea.
DNA extraction and PCR assays.
Genomic DNA (gDNA) was isolated from liquid cultures using standard protocols (28). Long-range PCR assays were performed in a final volume of 25 μl containing 20 to 50 ng of gDNA, 0.4 mM deoxynucleoside triphosphate (dNTP), 12.5 pmol each of the BAB1_0997F and BAB1_1008R primers (see Table S2 in the supplemental material), and 1 U of long-range enzyme mix (Fermentas Inc., Burlington, Canada). The PCR conditions used included an initial step at 94°C for 2 min, followed by 30 cycles at 94°C for 20 s, annealing at 60°C for 30 s, and extension at 68°C for 13 min. Finally, an extension step at 68°C for 10 min was performed. PCR products were identified by restriction analysis with PvuII (Fermentas). For chromosomal scar detection, PCR assays were performed in a final volume of 25 μl containing 0.2 to 0.5 μg of gDNA, 10 pmol each of BAB1_0981F and BAB1_1007R, 0.2 mM dNTPs, 2 mM MgCl2, and 1 U of Taq Platinum enzyme (Invitrogen, Carlsbad, CA). The mixture was preincubated at 95°C for 5 min, followed by 30 cycles at 95°C for 20 s, 60°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. The same conditions were used for circular intermediate (CI) detection using primer pair BAB1_1005F and BAB1_0983bR. All PCR were carried out in a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). Amplicons and restriction fragments were resolved by electrophoresis in 1.0 to 2.0% TAE (40 mM Tris-acetate, 1 mM EDTA; pH 8.0) agarose gels.
qPCR.
The frequency of detection of GI-2 deletion was assessed by absolute quantification using real-time PCR with gDNA of the ΔGI-2 strain (R1 strain) or plasmid DNA from the cloned 586-bp fragment carrying the attB site (see pGI2del in Table S1 in the supplemental material) to prepare calibration curves. Amplification rounds included 0.2 μg of gDNA, 0.3 μM each of the BAB1_0981F and BAB1_1007R primers (see Table S2 in the supplemental material), 4 mM MgCl2, 1 μl of LightCycler FastStart DNA SYBR green I master mix (Roche, Indianapolis, IN), and PCR-grade water to a final volume of 20 μl in a LightCycler1.5 real-time PCR system (Roche). The reaction was initiated by enzyme activation and DNA denaturation at 95°C for 10 min and 45 cycles at 95°C for 8 s, annealing at 65°C for 8 s, and extension at 72°C for 20 s. The specificity of the reaction was assessed by melting curve analysis using the LightCycler3 software. For this, the amplicons were heated to 95°C for 1 s and 70°C for 15 s and then heated slowly at 0.1°C/s to 99°C under continuous fluorescence monitoring. Each round included a negative control without DNA. The frequency of detection of GI-2 deletion was estimated from the ratio between the numbers of chromosomal scars and genome copies of the corresponding DNA sample assessed by A260 (16). All measurements were done in duplicate. Three independent experiments were performed with each strain.
Mutagenesis.
The construction of the B. abortus int mutant was accomplished by gene disruption (23). Briefly, primers BAB1_0983-mutF and -R (see Table S2 in the supplemental material) were used to amplify an internal fragment of int using B. abortus 2308 Nalr DNA as the template. This fragment was inserted into the BamHI-XbaI-digested pJQK plasmid, and the resulting pJQKint plasmid was transferred to the E. coli S17-1λpir strain and introduced into Brucella by mating. Integrative mutants were selected on nalidixic acid (25 μg/ml) and kanamycin (50 μg/ml). To confirm the site of the insertion, independent PCR rounds with primer pair BAB1_0983_F1 and M13R and primer pair BAB1_0983_R4 and M13R were performed (see Fig. S1 in the supplemental material). For complementation, a copy of int generated by PCR with primers BAB1_0983F-gw and BAB1_0983R-gw (see Table S2 in the supplemental material) was introduced in the plasmid vector pDONR221 using BP Clonase (Gateway technology; Invitrogen). The resulting plasmid pDONRint was used along with the pMR10 destination vector to obtain the complementation plasmid pMR10int by LR Clonase reaction and cloned into the E. coli OmniMAX 2-t1 strain. After this, the construct was introduced into the B. abortus int::Kmr mutant using the E. coli S17-1λpir procedure.
RNA extraction and reverse transcription assays.
Total RNA was prepared from 10 ml of B. abortus, B. melitensis, or B. suis cultures using RNeasy (Qiagen, Hilden, Germany). DNA contamination was removed by treatment with Turbo DNase (Ambion, Austin, Texas), and the yield was measured at A260. For cDNA synthesis, 100 ng of total RNA was reverse transcribed using 10 units of Transcriptor RT (Roche) and 0.3 μM BAB1_0983F-specific primer. The reaction mixture was incubated at 55°C for 30 min, as described by the manufacturer (Roche). A 401-bp internal fragment of int was amplified in a reaction mixture containing 2 μl of cDNA, 12.5 pmol of BAB1_0983F and -R primers, 0.4 mM dNTPs, 2 mM MgCl2, 1× PCR buffer, and 1 U of Taq Platinum (Invitrogen) with a final volume of 25 μl. Samples were incubated at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. At the end, a final extension step at 72°C for 5 min was included. Reverse transcription-PCR (RT-PCR) was performed in a GeneAmp 9700 thermal cycler (Applied Biosystems).
IS711 Southern blotting.
The IS711 sequence was identified by Southern blotting using 1 to 2 μg of AvaI-ClaI double-digested B. abortus gDNA. The product was resolved in 1% agarose at 15 mA for 10 h and probed with a biotin-labeled (BioNick; Invitrogen) IS711 fragment generated by PCR with primers 711u and 711d (15). Chemiluminescent detection of the hybridized product was done using a commercial kit (KPL, Gaithersburg, MD). Films were developed by conventional photographic methods.
Nucleotide sequence accession numbers.
Sequences of PCR products derived from the scar and the CI were deposited to GenBank under accession numbers EU128673 and EU128674, respectively.
RESULTS AND DISCUSSION
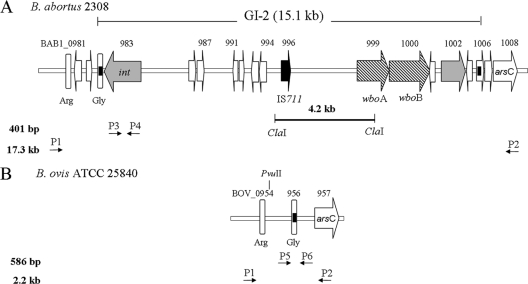
In B. abortus 2308, GI-2 is located in a 15.1-kb chromosomal region found between coordinates 953807 and 968958 of chromosome I (GenBank accession number AM040264) (5). This region complies with the structure of a canonical GI, as it is inserted into the tRNAGly gene sequence flanked by a pair of 41-bp identical direct repeats. GI-2 carries 13 open reading frames (ORFs), with an average of 51.3% of GC content. This is lower than the average of 57% for the whole genome, suggesting horizontal acquisition (7). A putative phage-related integrase (BAB1_0983) is located at the 5′ end of GI-2, and the two genes (wboA and wboB) encoding the O-polysaccharide glycosyltransferases are located at the 3′ end (Fig. 1A). B. abortus GI-2 shows 11 ORFs annotated as pseudogenes in comparison with the B. melitensis orthologs, as follows: a 25-kDa immunogenic protein precursor (BAB1_0989-990), a queuine tRNA ribosyltransferase (BAB1_0995), a phage-related protein (BAB1_0998), and the hypothetical proteins BAB1_0984-985, BAB1_0988, BAB1_0997, BAB1_0998, and BAB1_1004-1005. In silico restriction analysis of GI-2 predicted a 4.2-kb ClaI fragment carrying a copy of IS711 (Fig. 1A). In contrast to the S species, B. ovis lacks GI-2 (22, 25). Instead, there is a chromosomal scar with a putative attB site (Fig. 1B), suggesting that GI-2 excision may have occurred spontaneously and that this could be a cause of S-R dissociation in S Brucella species.
FIG. 1.
Structure of B. abortus GI-2 and the corresponding B. ovis locus. (A) Genetic organization of B. abortus 2308 15.1-kb GI-2. Gray arrows, phage-related genes (int, integrase gene); cross-hatched arrows, LPS biosynthesis genes; white arrows, unknown or hypothetical ORFs; black arrow, IS711 (containing the transposase ORF); white vertical bars, tRNA genes; black boxes (within the tRNA gene or a hypothetical ORF flanking the GI), direct repeats sequences. The positions of primers BAB1_0979F and BAB1_1008R (used to amplify a 17.3-kb fragment, including GI-2) are marked P1 and P2, and the positions of primers BAB1_0983F and -R (used to amplify a 401-bp fragment of the integrase) are marked P3 and P4, respectively. The ClaI-ClaI segment indicates the position of the corresponding IS711-carrying 4.2-kb restriction fragment. (B) Genetic organization of the B. ovis ATCC 25840 locus carrying the GI-2 excision scar. In this case, primers P1 and P2 amplify a 2.2-kb fragment spanning the attB-containing scar (black box) and a PvuII site. The positions of primers BAB1_0981F and BAB1_1007R (used to amplify a 586-bp internal fragment of the scar) are marked P5 and P6, respectively.
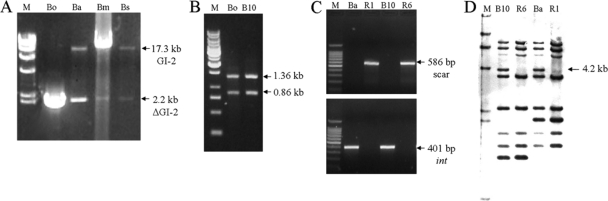
To assess the stability of GI-2, we grew B. abortus 2308 (S) (Table 1) in Brucella broth under standard conditions (37°C with continuous stirring) and performed long-range PCR for the detection of the complete GI-2 with primers BAB1_0979F and BAB1_1008R. Two amplicons were generated, one of approximately 17.3 kb that corresponded to the predicted product and a second one of 2.2 kb, which was similar to that amplified from B. ovis (Fig. 2A). These results suggest that a fraction of the cells in the B. abortus 2308 culture were undergoing the loss of GI-2. Then, we performed a similar experiment using B. abortus field isolate B10. Again, the same two types of PCR products were obtained (not shown). Moreover, restriction analysis of the B. abortus B10 2.2-kb PCR product showed a pattern similar to that obtained from B. ovis (Fig. 2B). We then selected two spontaneous R B. abortus mutants of different genetic backgrounds (R1 and R6) (Table 1) for further characterization. Like B. ovis, both mutants showed the attB site at the chromosomal scar and the absence of the GI-2 integrase (Fig. 2C). Sequencing of the product confirmed the identity of the scar, and the loss of GI-2 in the R1 and R6 chromosomes was further confirmed by the absence of a 4.2-kb IS711-containing fragment (Fig. 2D). Finally, we examined B. abortus, B. melitensis, and B. suis cultures for the presence of the circular intermediates (CIs) that should be released upon GI-2 excision (Fig. 3A). Consistent with this hypothesis, PCR targeted to the attP sites in the CIs using primers BAB1_1005F and BAB1_0983bR confirmed the release of GI-2 in B. abortus, B. melitensis, and B. suis (Fig. 3B). Moreover, sequence analysis showed the 41-bp repeat at the attP and flanking att sites, indicating the occurrence of site-specific recombination (Fig. 3C). All together, these results link the spontaneous excision of GI-2 to the appearance of the R phenotype in these three S Brucella species. We also examined the permanence of CIs in 5 spontaneous R mutants that carried the GI-2 deletion. None of these mutants were positive for CI detection by PCR, showing that the CIs had been lost beyond detection, consistent with the absence of a replication origin.
FIG. 2.
Excision of GI-2 occurs spontaneously. (A) Long-range PCR analysis (primers P1 and P2) (Fig. 1) for the presence of GI-2 in Brucella cultures. Bo, B. ovis; Ba, B. abortus 2308; Bm, B. melitensis 16M; Bs, B. suis 1330. Abbreviations: 17.3-kb GI-2, GI-2-containing fragment (Fig. 1); 2.2-kb ΔGI-2, chromosomal scar carrying fragment (Fig. 1); M, λ/HindIII DNA ladder (Fermentas). (B) PvuII restriction analysis of the 2.2-kb fragment of the following: B10, B. abortus B10, and Bo, B. ovis. M, 1-kb DNA ladder (Fermentas). (C) PCR analysis of R B. abortus mutants R1 and R6. (Top) Detection of the chromosomal scar with primers P5 and P6 (Fig. 1); (bottom) detection of the int gene with primers P3 and P4 (Fig. 1). M, 100-bp DNA ladder (Promega, Madison, WI). (D) IS711 Southern blot of the following: B10, B. abortus B10 (field isolate); R6, R B. abortus mutant derived from B10; Ba, B. abortus 2308; and R1, R B. abortus mutant derived from strain 2308. The arrow shows the position of the 4.2-kb ClaI restriction fragment (Fig. 1) that carries the IS711 copy. M, λ/HindIII biotin-labeled DNA ladder. All experiments were performed with DNA from broth cultures, except for 2C, in which DNA from a single bacterial colony was used.
FIG. 3.
GI-2 excision is promoted by the phage-related integrase gene. (A) Diagram of the excision intermediate showing the position of the 385-bp fragment carrying the recombined flanking GI-2 repeats (gray box), amplified by PCR with primers BAB1_1005F and BAB1_0983bR. (B) PCR detection of the 385-bp product amplified from the excision intermediate in weak acidic cultures of B. melitensis, B. suis, and B. abortus. M, 1-kb ladder (Roche). (C) Sequence alignment of the flanking att and attP sites. The 41-bp repeat is underlined. (D) Mutational analysis of the phage-related integrase. DNA from the B. abortus parental (2308 Nalr), mutant (int::Kmr), and complemented (int::Kmr/int) strains was used for PCR detection of the GI-2 scar (586 bp) and CI (385 bp).
It is known that low oxygen tension and acidic pH promote S-R Brucella dissociation and that R mutants tend to accumulate as cultures reach stationary phase (2, 4). By qPCR using the BAB1_0981F and BAB1_1007R primers, we estimated a GI deletion frequency of 1.0 ± 0.21 × 10−5 to 2.9 ± 0.32 × 10−5 for stationary-phase cells (Table 2). When we examined the dissociation of B. abortus 2308 using Brucella broth adjusted to pH 6.6 and prolonged the incubation for 10 days, we isolated R mutants with a frequency of 2.7 ± 0.25 × 10−2 (Table 2), with the GI-2 deletion accounting for only 10 to 20% of them. Moreover, although qPCR evidenced the occurrence of the GI-2 deletion, we did not detect the GI-2 chromosomal scar in any of the B. melitensis and B. suis R mutants isolated on plates. These results suggest that in addition to GI-2 excision, there are other mechanisms involved in S-R Brucella dissociation. One of these mechanisms could involve the transposition of insertion element (IS) sequences into GI-2, as suggested by the fact that the spontaneous R mutant RB51 presents IS711 inserted in wboA (24). Our results also leave open the possibility that such alternative mechanisms of S-R dissociation could have different relevance in different Brucella strains or species. This is suggested by the different estimated frequencies observed by qPCR in B. melitensis 16M, B. suis 1330, and B. abortus 2308 grown under acidic conditions (Table 2). Research is in progress to study these possibilities.
TABLE 2.
Frequencies of S-R dissociation and of detection of the GI-2 deletion
| Strain | Growth conditions | Dissociation frequency (10−2)a | Frequency of detection of GI-2 deletion by: |
|
|---|---|---|---|---|
| PCR (10−3)b | qPCR (10−5)c | |||
| B. melitensis 16M | 37°C, pH 6.6, 10 days | 0.1 ± 0.03 | Not detected | 10 ± 3 |
| B. suis 1330 | 37°C, pH 6.6, 10 days | 0.7 ± 0.10 | Not detected | 67 ± 15 |
| B. abortus 2308 | 37°C, pH 6.6, 10 days | 2.7 ± 0.25 | 4.5 ± 1.1 | 900 ± 210 |
| B. melitensis 16M | 37°C, late-stationary phase | Not detected | Not detected | 1.0 ± 0.21 |
| B. suis 1330 | 37°C, late-stationary phase | Not detected | Not detected | 1.3 ± 0.25 |
| B. abortus 2308 | 37°C, late-stationary phase | Not detected | Not detected | 2.9 ± 0.32 |
R colonies were identified by using the crystal violet dye exclusion test, and the dissociation frequency was calculated with respect to the total colony count.
PCR detection of the chromosomal scar was carried out for each R colony, and then the GI-2 deletion frequency was assessed (values represent the means and standard errors of the means of results from three independent experiments).
Values represent the means and standard errors of the means of results from three independent experiments. The frequency of detection of the GI-2 deletion was estimated as the ratio between the numbers of scars and genome copies. The efficiency (E) of the PCR obtained within the range of 106 to 102 target copies was 1.84 ± 0.07.
We registered the expression of the putative integrase/recombinase (BAB1_0983) gene in all cases tested (data not shown), and we hypothesized that it could be involved in the excision. Accordingly, we constructed a B. abortus 2308 Nalr int::Kmr mutant by gene disruption (see Fig. S1 in the supplemental material) and the corresponding int::Kmr/int-complemented derivative. By PCR, we identified both the chromosomal scar and the CIs from cultures of the parental strain and in the int::Kmr/int-complemented strain but not in the mutant (Fig. 3D). In addition, the assessment of the S-R dissociation rates of B. abortus 2308 Nalr and B. abortus 2308 int::Kmr demonstrated a stabilization of the S phenotype associated to the int mutation (Fig. 4).
FIG. 4.
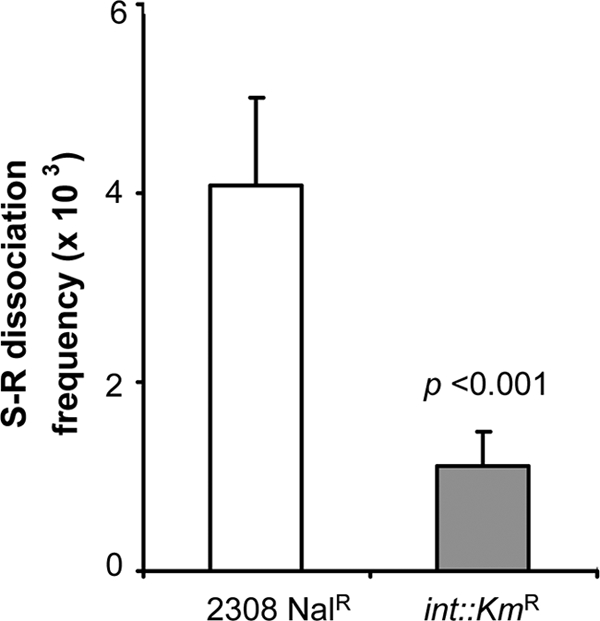
Dissociation frequencies of the parental strain of B. abortus (2308 Nalr) and the mutant in the phage-related integrase gene (int::Kmr). The means and standard errors of the means of the results from three independent experiments were 4.1 ± 0.9 × 10−3 and 1.1 ± 0.3 × 10−3, respectively (P < 0.001; chi-square test).
Finally, it is worth commenting on the role of the S-R dissociation in the biology of the genus Brucella. It has been proposed that GI circularization facilitates its transfer within a bacterial population, thus enhancing the genetic adaptability and competitiveness during host-pathogen interactions (19). Also, it has been suggested that GI deletion is accompanied by the appearance of a less virulent phenotype, a result that could help some bacteria to succeed in long-term colonization during infection (10). However, these hypotheses are unlikely to be relevant in Brucella. First, this pathogen shows high clonality, with no evidence of any mechanism of gene exchange at the population level, in keeping with its intracellular lifestyle (6). Second, it has been a consistent observation that spontaneous and laboratory-produced R Brucella mutants are severely attenuated in cells and animals (1, 21), making it unlikely that GI-2 deletion plays a role in the perpetuation of the disease. However, this does not mean that the excision does not happen within the host. In fact, the GI-2 chromosomal scar has been identified in DNA obtained from abomasum fluids of B. abortus-infected cattle (M. Mancilla, unpublished results), and one possibility is that the acidic environment of the abomasum might have facilitated GI-2 excision. The significance of the S-R dissociation in this and possibly other extracellular fluids is unclear. R Brucella mutants are also occasionally isolated from the milk of infected goats (C.M. Marín, J. M. Blasco, and I. Moriyón, unpublished observations), and they may represent variants that, although able to multiply in environments less stringent than the intracellular niche, are not transmitted from host to host. GI-2 instability may have played a role in speciation within the genus Brucella. Even though B. ovis and B. canis carry the major O-polysaccharide wbk genetic region, both are naturally rough (R) (8). Previous works have shown that B. ovis carries a frameshift in wbkF (29) and a GI-2 deletion (25), and here we provide evidence for the mechanism behind this deletion. On the other hand, B. canis conserves GI-2, and its R phenotype has been traced to a deletion overlapping wbkD and wbkF (29). These data show that the emergence of B. canis and B. ovis has followed different paths, consistent with the clustering of the former with B. suis, and the evidence suggests that B. ovis forms a more ancient lineage (12, 26).
Supplementary Material
Acknowledgments
We thank Minie Villarroel from Servicio Agrícola y Ganadero de Chile for maintaining and providing the Brucella strains.
This work was funded by the Department of Research and Development, Universidad Austral de Chile, project S-2009-33, FONDEF D02I-1111, Ministerio de Ciencia y Tecnología of Spain (AGL2008-04514), and by EU grant agreement 221948 (“Integrated Control of Neglected Zoonoses: Improving Human Health and Animal and Animal Production through Scientific Innovation and Public Engagement”). M.M. was supported by the CONICYT-Chile fellowship and PIUNA University of Navarra.
Footnotes
Published ahead of print on 15 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allen, C. A., L. G. Adams, and T. A. Ficht. 1998. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 66:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altenbern, R., D. Williams, J. Kelsh, and W. Mauzy. 1956. Metabolism and population changes in Brucella abortus II: terminal oxidation and oxygen tension in population changes. J. Bacteriol. 73:697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alton, G., L. Jones, R. Angus, and J. M. Verger (ed.). 1988. The production of Brucella vaccines. Techniques for the brucellosis laboratory. INRA, Paris, France.
- 4.Braun, W. 1946. Dissociation in Brucella abortus: a demonstration of the role of inherent and environmental factors in bacterial variation. J. Bacteriol. 51:327-349. [DOI] [PubMed] [Google Scholar]
- 5.Chain, P. S., D. J. Comerci, M. E. Tolmasky, F. W. Larimer, S. A. Malfatti, L. M. Vergez, F. Aguero, M. L. Land, R. A. Ugalde, and E. Garcia. 2005. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect. Immun. 73:8353-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ficht, T. A. 2003. Intracellular survival of Brucella: defining the link with persistence. Vet. Microbiol. 92:213-223. [DOI] [PubMed] [Google Scholar]
- 7.Gal-Mor, O., and B. B. Finlay. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell. Microbiol. 8:1707-1719. [DOI] [PubMed] [Google Scholar]
- 8.Godfroid, F., A. Cloeckaert, B. Taminiau, I. Danese, A. Tibor, X. de Bolle, P. Mertens, and J. J. Letesson. 2000. Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151:655-668. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez, D., M. J. Grillo, M. J. De Miguel, T. Ali, V. Arce-Gorvel, R. M. Delrue, R. Conde-Alvarez, P. Munoz, I. Lopez-Goni, M. Iriarte, C. M. Marin, A. Weintraub, G. Widmalm, M. Zygmunt, J. J. Letesson, J. P. Gorvel, J. M. Blasco, and I. Moriyon. 2008. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS One 3:e2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacker, J., U. Hentschel, and U. Dobrindt. 2003. Prokaryotic chromosomes and disease. Science 301:790-793. [DOI] [PubMed] [Google Scholar]
- 11.Mancilla, M., M. Villarroel, M. E. Saldías, J. Soto, and A. M. Zárraga. 2008. Genotipos de aislados de campo de Brucella abortus de distintas regiones geográficas de Chile. Arch. Med. Vet. 40:187-192. [Google Scholar]
- 12.Moreno, E., A. Cloeckaert, and I. Moriyon. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90:209-227. [DOI] [PubMed] [Google Scholar]
- 13.Moreno, E., and I. Moriyon. 2007. The genus Brucella, p. 315-455. In S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 5. Springer-Verlag, New York, NY. [Google Scholar]
- 14.Moriyon, I., M. J. Grillo, D. Monreal, D. Gonzalez, C. Marin, I. Lopez-Goni, R. C. Mainar-Jaime, E. Moreno, and J. M. Blasco. 2004. Rough vaccines in animal brucellosis: structural and genetic basis and present status. Vet. Res. 35:1-38. [DOI] [PubMed] [Google Scholar]
- 15.Ocampo-Sosa, A. A., J. Aguero-Balbin, and J. M. Garcia-Lobo. 2005. Development of a new PCR assay to identify Brucella abortus biovars 5, 6 and 9 and the new subgroup 3b of biovar 3. Vet. Microbiol. 110:41-51. [DOI] [PubMed] [Google Scholar]
- 16.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guedon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759-774. [DOI] [PubMed] [Google Scholar]
- 17.Rajashekara, G., J. Covert, E. Peterson, L. Eskra, and G. Splitter. 2008. Genomic island 2 of Brucella melitensis is a major virulence determinant: functional analyses of genomic islands. J. Bacteriol. 190:6243-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajashekara, G., J. D. Glasner, D. A. Glover, and G. A. Splitter. 2004. Comparative whole-genome hybridization reveals genomic islands in Brucella species. J. Bacteriol. 186:5040-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon, R., U. Priefer, and A. Pehle. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-890. [Google Scholar]
- 21.Spink, W. 1956. The nature of brucellosis, p. 53-58. The University of Minnesota Press, Minneapolis, MN.
- 22.Tsolis, R. M., R. Seshadri, R. L. Santos, F. J. Sangari, J. M. Lobo, M. F. de Jong, Q. Ren, G. Myers, L. M. Brinkac, W. C. Nelson, R. T. Deboy, S. Angiuoli, H. Khouri, G. Dimitrov, J. R. Robinson, S. Mulligan, R. L. Walker, P. E. Elzer, K. A. Hassan, and I. T. Paulsen. 2009. Genome degradation in Brucella ovis corresponds with narrowing of its host range and tissue tropism. PLoS One 4:e5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uzureau, S., M. Godefroid, C. Deschamps, J. Lemaire, X. De Bolle, and J. J. Letesson. 2007. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 189:6035-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vemulapalli, R., J. R. McQuiston, G. G. Schurig, N. Sriranganathan, S. M. Halling, and S. M. Boyle. 1999. Identification of an IS711 element interrupting the wboA gene of Brucella abortus vaccine strain RB51 and a PCR assay to distinguish strain RB51 from other Brucella species and strains. Clin. Diagn. Lab Immunol. 6:760-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vizcaino, N., P. Caro-Hernandez, A. Cloeckaert, and L. Fernandez-Lago. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6:821-834. [DOI] [PubMed] [Google Scholar]
- 26.Whatmore, A. M. 2009. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9:1168-1184. [DOI] [PubMed] [Google Scholar]
- 27.White, P. G., and J. B. Wilson. 1951. Differentiation of smooth and nonsmooth colonies of brucellae. J. Bacteriol. 61:239-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, K. 2001. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. Chapter 2:Unit 2.4. [DOI] [PubMed]
- 29.Zygmunt, M. S., J. M. Blasco, J. J. Letesson, A. Cloeckaert, and I. Moriyon. 2009. DNA polymorphism analysis of Brucella lipopolysaccharide genes reveals marked differences in O-polysaccharide biosynthetic genes between smooth and rough Brucella species and novel species-specific markers. BMC Microbiol. 9:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.