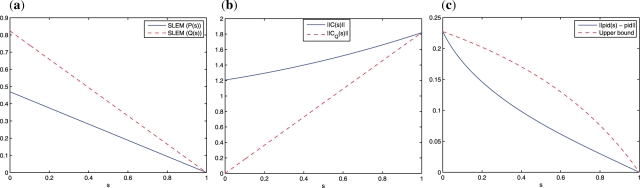
Fig. 3.
The human melanoma gene regulatory network: (a) Plot of SLEM(P(s)) in Equation (17) (blue continuous line) and SLEM(Q(s)) in Equation (22) (red dashed line) versus s; (b) Plot of ‖C(s)‖ in Equation (16) (blue continuous line) and ‖CQ(s)‖ in Equation (21) (red dashed line) versus s; (c) Plot of ‖πd(s) − πd‖ (blue continuous line), and the upper bound 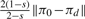 (red dashed line) versus 0 ≤ s ≤ 1, in Proposition 5. The trade-off between minimal-energy and fastest convergence rate control is clear from (a) and (b). The parameterized family of perturbed matrices P(s) in Equation (15) results in a faster convergence toward the desired steady-state πd at the expense of a higher norm (energy) of the perturbation matrix. On the other hand, the family of perturbed matrices Q(s) in Equation (20) leads to a perturbation matrix with norm (energy) as small as desired, but at the expense of not converging toward the desired steady state for small s. The distance between the steady state of Q(s) and the desired steady state as a function of s is shown in (c).
(red dashed line) versus 0 ≤ s ≤ 1, in Proposition 5. The trade-off between minimal-energy and fastest convergence rate control is clear from (a) and (b). The parameterized family of perturbed matrices P(s) in Equation (15) results in a faster convergence toward the desired steady-state πd at the expense of a higher norm (energy) of the perturbation matrix. On the other hand, the family of perturbed matrices Q(s) in Equation (20) leads to a perturbation matrix with norm (energy) as small as desired, but at the expense of not converging toward the desired steady state for small s. The distance between the steady state of Q(s) and the desired steady state as a function of s is shown in (c).