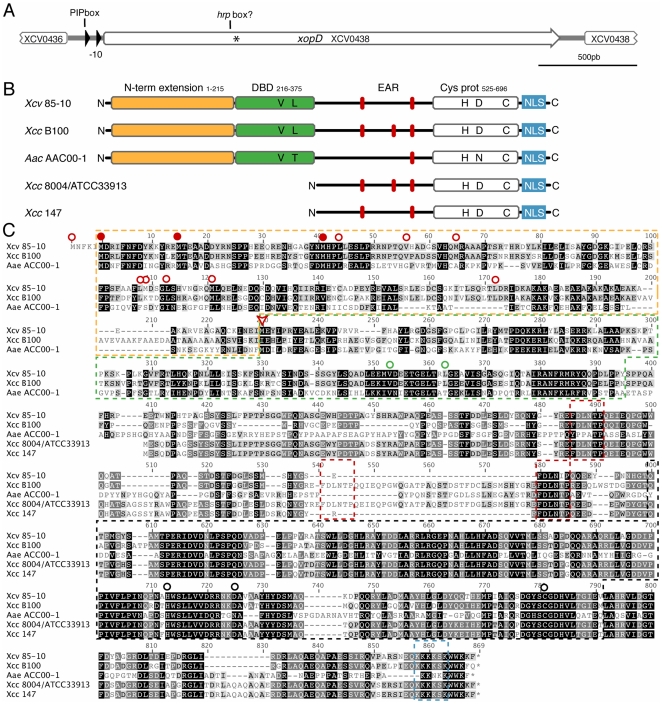
Figure 1. In silico analysis of the xopD locus.
(A) The xopD locus in Xcv 85-10 is shown (the genome interval for the Xcv 85-10 shown sequence and the xopD gene are respectively: 486209–489200 and 486544–488826). Open arrows indicate ORFs and filled arrows show promoter elements, such as the PIP box and the −10 sequence. The position of XopD translation start annotated in the public databases is indicated by an asterisk. (B) Schematic representation of XopD functional domains in Xcv 85-10, in comparison to its known protein homologues. The N-terminal extension, essential V and L residues in the DBD, tandemly repeated EAR motifs, conserved catalytic residues in the cysteine protease domain, and NLS motif are shown. (C) Sequence alignment of XopD from Xcv 85-10, hypothetical protein from Xcc B100 (YP_001902662), virulence protein from Xcc 8004 (AAY48282) or Xcc ATCC 33913 (AAM42168), peptidase C48 SUMO from Acidovorax avenae (EFA39722) and XopD from Xcc 147. The figure shows the longest ORF possible for all proteins. For XopD from Xcv, the first shown M residue corresponds to a UUG codon, not conserved among Xcc B100 and A. avenae coding sequences. The M residue previously annotated as the starting amino acid of the XopD protein is indicated by a red arrowhead. Putative translation starts situated in frame and upstream the previously annotated starting M are also indicated as follows: translation starts conserved among Xcv 85-10, Xcc B100 and Aea are indicated with a full red dot, otherwise, they appear indicated by an empty red dot. Yellow box: putative N-terminal extension; green box: DNA-binding domain (V and L residues in the helix-loop-helix domain essential for maximal DNA-binding [46] are indicated by an empty green dot); red boxes: tandemly-repeated EAR motifs [L/FDNLL/F(X)P] [44]; black box: SUMO protease domain (H, D and C catalytic core residues are indicated by a black empty dot); blue box: NLS. Gray and black highlighting of amino acids indicates, respectively, 70–80% and 90–100% of similarity.