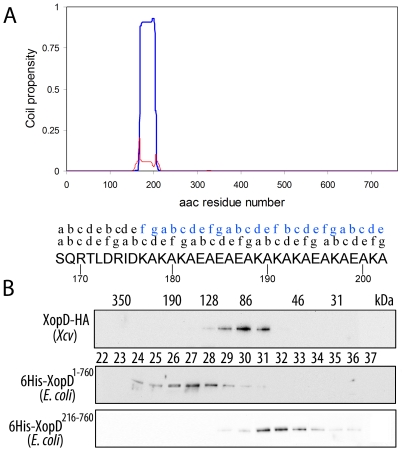
Figure 4. XopD1-760, but not XopD216-760, is able to dimerize.
(A) Multicoil (http://groups.csail.mit.edu/cb/multicoil/cgi-bin/multicoil.cgi) prediction on XopD1-760 sequence. XopD amino acid sequence (amino acids 1 to 760) was analyzed for its propensity to form coiled-coil structures. A region comprising amino acid residues 168–202 showed high probability to form such coiled-coil structures. Furthermore, this coiled-coil region is likely to be involved in dimer formation (blue line), rather than trimer (red line) or other oligomeric structures. The (abcdefg) positions of the hepta-repeat in the coiled-coil region are indicated above the amino acid sequence. Above it, the relative positions of the hepta-repeat in the opposite chain within the putative dimer are indicated in blue. (B) Gel filtration analysis of HA-tagged XopD expressed in Xcv, and XopD1-760 and XopD216-760 expressed in E. coli as 6xHis-tagged fusions. Protein extracts were subjected to gel filtration chromatography on a Superdex S-200 column. 0.4 ml fractions were collected and aliquots were analyzed by Western blot. Fraction numbers of the elution profile re indicated by the numbers between the gels. The molecular mass estimated for each fraction (in kDa) is given at the top.