Figure 4.
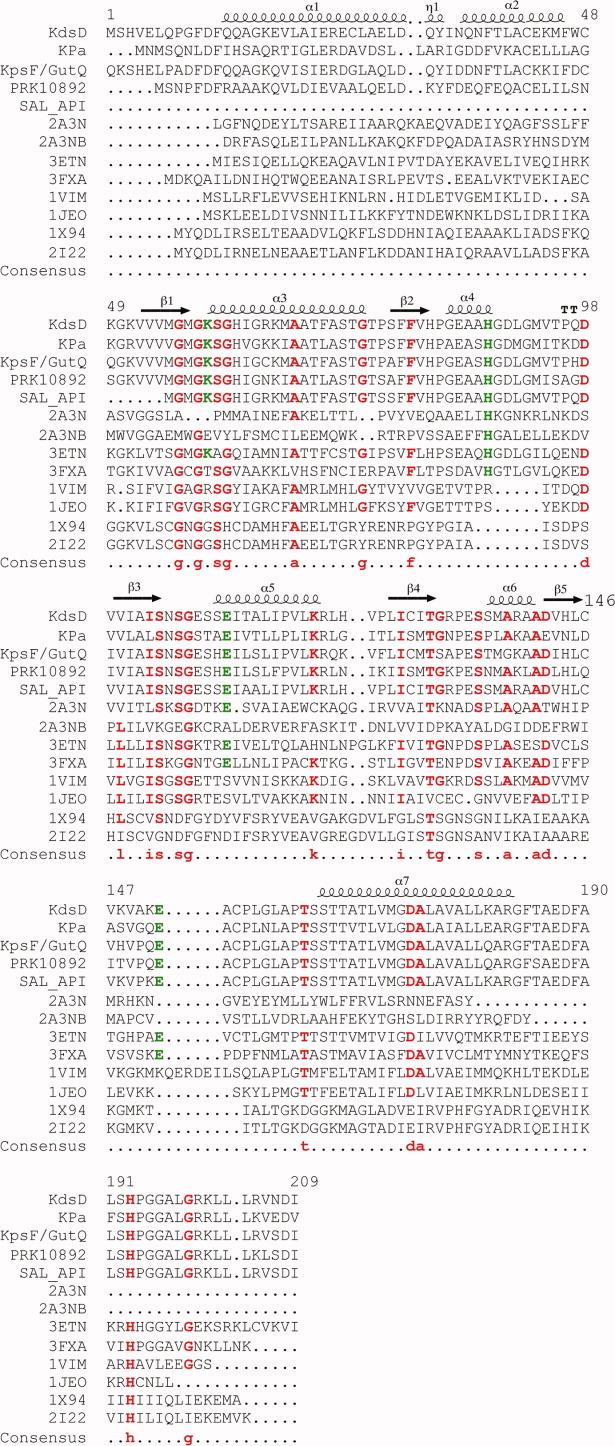
Multiple sequence alignments of SIS domains. Residues 1–209 of KdsD were aligned against single-domain (PDB entries 1VIM, 1JEO, 2I22, 3ETN, 3FXA, and 1X94) and the two-domain (2A3N) phosphosugar isomerase of known structure, and four, two-domain proteins (KdsD from Pseudomonas aeruginosa, KpsF/GutQ family protein from Pectobacterium carotovorum, d-arabinose-5-phosphate from Salmonella enterica, and a putative polysialic acid capsule expression protein from Vibrio parahaemolyticus), of unknown structure, alongside the secondary structure arrangement of KdsDK59A (residues 10–183). The sequences of the two diverse SIS domains of 2A3N are shown covering residues 1–171 (2A3N) and 172–343 (2A3NB). Consensus amino acids, including H88 and H193, and residues (K59, E111, and E152) known to be catalytically important in KdsDK59A are indicated in red and green font, respectively. Multiple sequence and secondary structure alignments were generated using the program MultAlin (http://multalin.toulouse.inra.fr/multalin/) and ENDscript1.1.
