Figure 4. Southern, genotyping and RT-PCR assays to characterize the Noxa1 conditional allele.
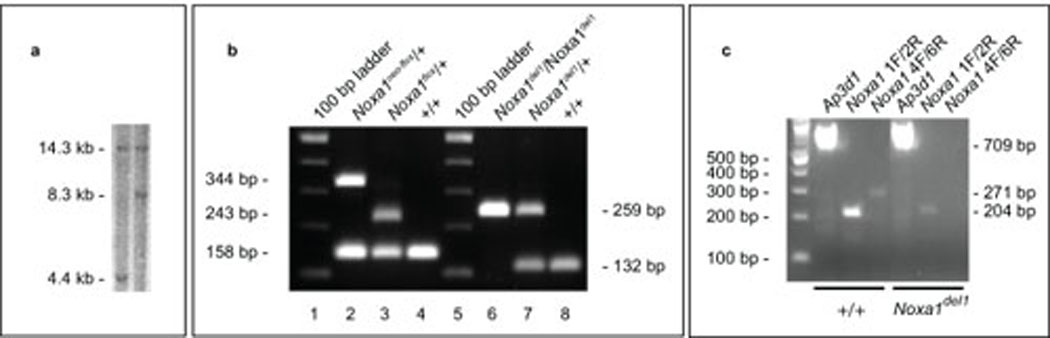
a. Southern analysis of targeted ES cell colonies identifies clones showing retention (left lane) or loss (right lane) of the loxP site in intron 2. b. Genotyping assays. Lanes 1–4 and lanes 5–8 showing the amplification products of two genotyping assays used to distinguish among the Noxa1+, Noxa1neo-flox, Noxa1flox and Noxa1del1 alleles (for details see Methods). c. RT-PCR analysis of Noxa1 transcripts from +/+ and Noxa1del1/Noxa1del1 temporal bone/inner ear RNA. Amplification of cDNA sequences upstream of the Cre-mediated deletion (primers 1F and 2R) is reduced in Noxa1del1 homozygotes. Amplification of cDNA sequences within the Cre-mediated deletion (primers 4F and 6R) is absent in Noxa1del1 homozygotes. Amplification of cDNA sequences from Ap3d1 serves as a control for the quality of the RNA.
