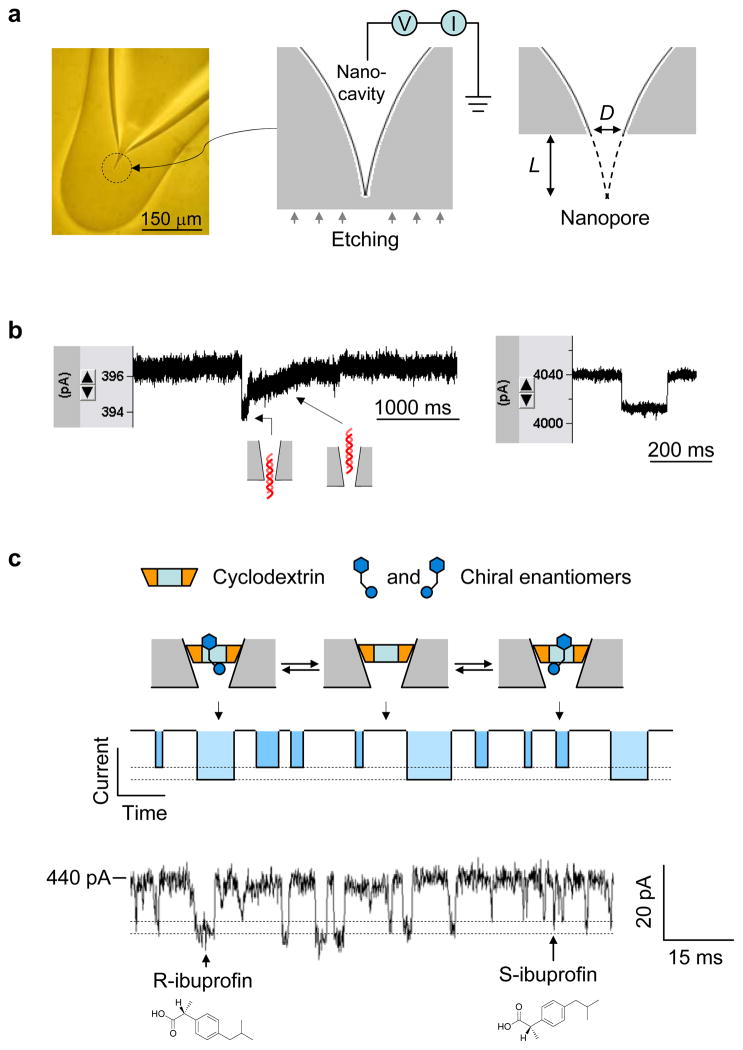
Figure 7.
Glass nanopore-terminated probe and single molecule manipulation. (a) The glass nanopore is fabricated by sealing the micropipette terminal to enclose a nanocavity, followed by external etching the glass terminal with electrical monitoring to perforate the nanocavity with controllable pore size. (b) Current blocks showing translocation of 1 kbp dsDNA through a 2 nm nanopore (left) and a 7 nm pore (right) in 1 M NaCl and recorded +100 mV. When the pore size is comparable to dsDNA, the DNA translocation speed is slowed down and translocation steps can be revealed from the block type (left), which is different from that for DNA translocation in a wider pore (right). (c) Single molecule discrimination of chiral enantiomers by the cyclodextrin (1.5 nm) trapped in the 1 nm nanopore. The interaction of chiral compounds with cyclodextrin can be separated from their block durations and current amplitudes. The current trace showed the binding of individual enantiomers of ibuprofen to the trapped β-cyclodextrin in the nanopore. The solution contained the mixture of 100 μM R-(−)-ibuprofen and 100 μM S-(+)-ibuprofen. The current block levels by R- and S-ibuprofen were marked with dash lines25.