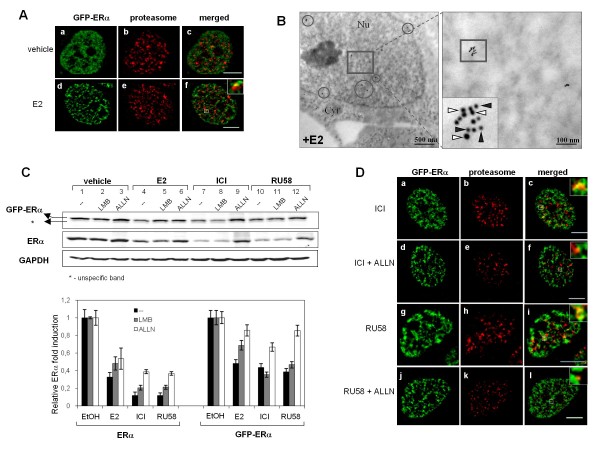
Figure 4.
Nuclear Proteasome-GFP-ERα contacts are frequent in the presence of E2 and SERDs. A) SK19 cells treated for 3 h with drugs as described in Methods. Cells were fixed and subjected to immunofluorescence using a monoclonal anti-20S proteasome subunit α2 primary antibody, followed by incubation with the Alexa Fluor® 647 goat anti-mouse secondary antibody. Right panels represent colocalization of GFP-ERα and 20S proteasome (insets): untreated cells (4A,c) and 10nM E2 treated cells (4A,f),. Images are representative of three independent experiments. Bar, 5 μm. B) 80 nm thin sections of MCF-7 cells were incubated with anti-ERα and anti-20S proteasome subunit α2 antibodies. Secondary antibodies coupled with 10 nm and 6 nm gold particles were used against anti-ERα (white arrowhead) and anti-20S proteasome (black arrowhead), respectively. Grids were then processed for electron microscopy. Colocalization of both proteins is indicated by circles. Bar, 500 nm. The area of selected electron micrograph shows colocalization clusters where at least 3 gold particles for each protein are present (inset). Bar, 100 nm. C) Western blot showing SERD-mediated degradation of ERα through the nuclear proteasome. SK19 cells were pre-treated or not with inhibitor of nuclear export (10 nM LMB) or proteasome inhibitor (100 μM ALLN) for 30 min and then treated with vehicle (EtOH as reference), 10 nM E2, 1 μM ICI, or 1 μM RU58 for 3 h. ERα and GAPDH (internal control) detection by Western blotting shows proteasome-dependent degradation of ERα upon E2 and SERDs stimulation. D) SK19 cells treated for 3 h with SERDs were pre-treated or not with proteasome inhibitor ALLN (details see in Methods). Right panels represent colocalization of GFP-EPα and 20S proteasome subunit α2 (insets): SERDs pre-treated cells without (4Dc and i) or with (4Df and l) 100 μM ALLN. Images are representative of three independent experiments. Bar, 5 μm.