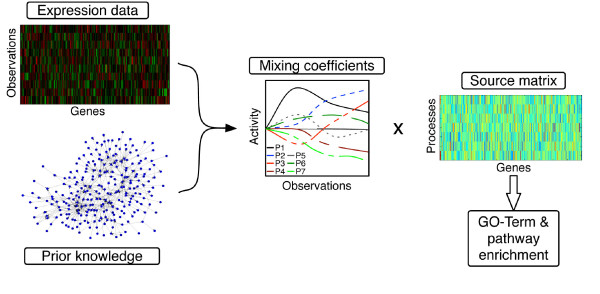
Figure 1.
GraDe: Graph-decorrelation algorithm. In cells, various biological processes are taking place simultaneously. Each of these processes has its own characteristic gene expression pattern, but different processes may overlap. A cell's total gene expression is then the sum of the expression patterns of all active processes, weighted by their current activation level. The GraDe algorithm combines a matrix factorization approach with prior knowledge in form of an underlying regulatory network. The input of GraDe is the transcriptional expression data, where observations can be different conditions or a time points, and the underlying regulatory network (prior knowledge). GraDe decomposes the observed expression data into the underlying sources S and their mixing coefficients A. Analyzing time-course microarray data, we interpret these sources as the biological processes and the mixing coefficients as their time-dependent activities. Observations indicate their expression behavior either in the different conditions or time-points and activity their activation strength. We further filter process-related genes by taking only the genes with the strongest contribution in each process. Finally, we test for enrichment of cellular processes (GO) and biological pathways (KEGG).