Abstract
Every polyketide synthase module has an acyl carrier protein (ACP) and a ketosynthase (KS) domain that collaborate to catalyze chain elongation. The same ACP then engages the KS domain of the next module to facilitate chain transfer. Understanding the mechanism for this orderly progress of the growing polyketide chain represents a fundamental challenge in assembly line enzymology. Using both experimental and computational approaches, the molecular basis for KS–ACP interactions in the 6-deoxyerythronolide B synthase has been decoded. Surprisingly, KS–ACP recognition is controlled at different interfaces during chain elongation versus chain transfer. In fact, chain elongation is controlled at a docking site remote from the catalytic center. Not only do our findings reveal a new principle in the modular control of polyketide antibiotic biosynthesis, they also provide a rationale for the mandatory homodimeric structure of polyketide synthases, in contrast to the monomeric nonribosomal peptide synthetases.
Keywords: assembly line enzymes, polyketide synthase, protein–protein interactions
Modular polyketide synthases such as 6-deoxyerythronolide B synthase (DEBS) are assembly lines of catalytic modules responsible for the biosynthesis of polyketide natural products (1–6). A fundamental challenge toward elucidating their molecular logic is to understand the mechanism by which covalently bound biosynthetic intermediates are sequentially accessed by individual catalytic sites of the synthase. Most notably, each module of a polyketide synthase (PKS) has an acyl carrier protein (ACP) domain that collaborates with the β-ketosynthase (KS) domain of the same module to catalyze polyketide chain elongation, and subsequently engages with the KS domain of the next module to facilitate forward chain transfer (Fig. 1). The simplest explanation for the orderly progress of a growing polyketide chain along a multimodular PKS is that each ACP is equivalently recognized by both KS partners, and that selection of the appropriate KS–ACP pair at a given point in the catalytic cycle is solely dictated by the identity of the substrate anchored on the ACP. (In the chain elongation reaction, a nucleophilic malonyl extender unit is attached to the ACP, whereas the growing polyketide chain itself is attached to the ACP via an electrophilic thioester linkage when it is ready for forward transfer.) Previous studies, however, suggest that protein–protein recognition between the KS and the ACP domains also plays an important role in both reactions (7–10). Moreover, this protein–protein recognition not only influences the specificity (kcat/KM) of each reaction, but also the maximum rate constant (kcat).
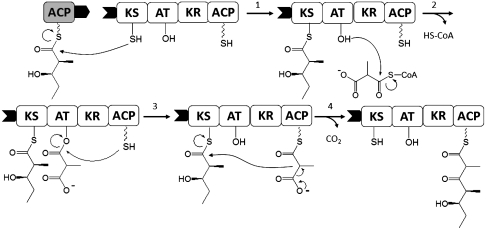
Fig. 1.
Chain elongation cycle catalyzed by a PKS module. Each module in a PKS consists of a unique set of covalently linked catalytic domains responsible for a single round of chain elongation via the following steps. (1) KS is primed with a growing polyketide chain by the upstream ACP (grayscale). AT is acylated with a CoA derived methylmalonyl extender unit (2), which is transferred to the downstream ACP (black and white) (3) and a decarboxylative condensation takes place in the active site of KS (4). The ACP tethered extended polyketide chain is processed by the tailoring domains (for example KR, process not shown) and subsequently transferred to the downstream KS domain as in (1). The phosphopantetheine prosthetic group of the ACP is drawn as a wavy line. Interprotein linkers that are known to facilitate intermodular chain transfer are shown as matching solid tabs (black). KS, β-ketosynthase; AT, acyltransferase; ACP, acyl carrier protein; KR, ketoreductase.
In the course of our efforts to elucidate the principles that govern KS–ACP recognition during chain elongation and chain transfer, we have made two unexpected and potentially important observations. First, notwithstanding the fact that the same active sites are deployed in both reactions, the structural elements that mediate KS–ACP recognition are entirely different in the two types of encounters. Second, the specificity of chain elongation is not controlled at the interface between the reactive KS–ACP pair. Rather, it is primarily dictated by interactions between the ACP and the subunit of the homodimeric module that is not involved in this catalytic event. Thus, a functional rationale has emerged for the universally conserved homodimeric architecture of PKS modules.
Results and Discussion
Site-Directed Mutagenesis of Helix II of the ACP.
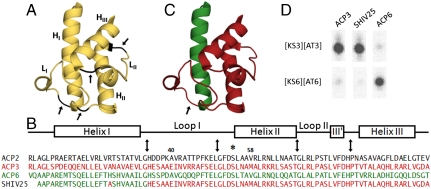
Numerous studies have focused on the protein recognition features of the ACP domains in fatty acid synthases and polyketide synthases (PKSs) (11–18). For example, helix II of the ACP was identified as the determinant of specificity between an ACP domain of DEBS and phosphopantetheinyl transferases that prime the ACP with a pantetheinyl “swinging arm” (13). Based on an analysis of the electrostatic features of the ACP domains of DEBS, helix II was also identified as potentially important in KS–ACP interactions (Fig. 2A) (19). Specifically, it was proposed that several charged residues on helix II might enable ACP3 and ACP6 to differentiate between their partner KS domains. Site-directed mutagenesis, however, failed to provide support for this model. Several single or double mutants of the charged residues on helix II of the ACP3 (K62N, R63Q, R71A, K62N/R71A, and R63Q/R71A) were prepared and analyzed. All mutants displayed identical specificity for KS3 over KS6 as wild-type ACP3 (Fig. S1). We therefore turned to the construction and analysis of chimeric ACPs to elucidate the molecular basis for orthogonal recognition within individual DEBS KS3–ACP3 and KS6–ACP6 pairs during polyketide chain elongation (9).
Fig. 2.
Structure-based hybrid ACP construction and analysis. (A) Solution structure of the ACP domain from DEBS module 2. To recombine the predicted secondary structure elements of ACP3 and ACP6 (helices HI, HII, and HIII, and loops LI and LII), four fusion sites marked by black arrows were identified. (B) Sequence alignment of DEBS ACP2, ACP3, ACP6, and a representative chimeric protein [construct SHIV25, whose model is shown in (C)]. In SHIV25 the N-terminal sequence of ACP6 is fused to the C-terminus of ACP3 at the loop I—helix I junction. The asterisk indicates the conserved phosphopantetheine attachment site Ser54. The arrows mark the fusion sites. Other fusion sites used to generate chimeras of ACP3 and ACP6 detailed in Fig. 3 are indicated by residue number. Red: ACP3-derived. Green: ACP6-derived. (D) Chain elongation activity of ACP3, ACP6, and the chimeric ACP SHIV25. Each ACP was assayed separately with [KS3][AT3] and [KS6][AT6] under conditions described in Materials and Methods section. The reaction product was visualized and quantified by radio-TLC.
Design of Chimeric ACPs.
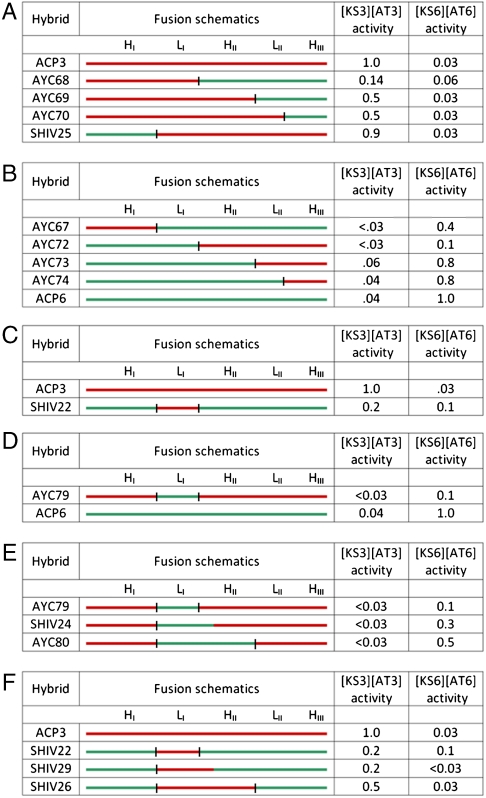
Based on the solution structure of DEBS ACP2, four junction sites between secondary structural elements— helix I (HI)—loop I (LI), loop I (LI)—helix II (HII), helix II (HII)—loop II (LII), and loop II (LII)—helix III (HIII)—were identified (Fig. 2). An initial set of 8 chimeras of ACP3 and ACP6 were prepared at each of these junctions (AYC67-AYC70, SHIV25 and AYC72-AYC74, Fig. 3 A and B). For steric reasons, an additional H26A mutation was also engineered into all chimeras (e.g., SHIV25) harboring both helix I from ACP6 and loop I from ACP3 (SI Appendix).
Fig. 3.
Evaluation of chimeric ACP proteins in an assay for polyketide chain elongation. A series of chimeric ACP proteins were constructed and assayed with homodimeric [KS][AT] proteins harboring either KS3 or KS6. For each chimera the activity with [KS3][AT3] or [KS6][AT6] is normalized to the corresponding wild-type ACP. The initial set of chimeras are shown in panels A and B. (A) ACPs that prefer [KS3][AT3] over [KS][AT6]. (B) ACPs that prefer [KS6][AT6] over [KS3][AT3]. Because these preferences tracked with the identity of LI, SHIV22 (C) and AYC79 (D) were constructed and found to have the predicted [KS][AT] preference, albeit at the expense of reduced activity. Additional chimeras, shown in Panels E and F, were engineered to further optimize activity and specificity. Specifically, redefinition of the LI-HII junction of SHIV22 and AYC79 afforded SHIV24 (E) and SHIV29 (F), respectively. The color scheme is similar to Fig. 2 (red = ACP3 derived, green = ACP6 derived). Fusion sites (black bars) are consistent with Fig. 2. For SHIV24 and SHIV29 the C-terminus of the substituted fragment is at residue 58. SHIV22, 25, 26 and 29 harbor a H26A mutation (see SI Text for details).
LI Is the Principal Determinant of KS–ACP Recognition During Polyketide Chain Elongation.
To compare quantitatively the ability of KS3 and KS6 to interact with each chimeric ACP, chain elongation was assayed with 190 kD homodimeric [KS][AT] didomain fragments of the corresponding DEBS modules (Fig. S2A). These structurally characterized truncated modules included KS and AT domains together with both flanking and intervening linker sequences but lacked the KR and ACP domains of the parent DEBS modules (9, 10, 20). Although this assay involves several elementary steps (for details, see Fig. S2), earlier studies have established that, under our assay conditions, KS–ACP catalyzed chain elongation (i.e., the docking of the methylmalonyl-charged ACP to the KS followed by C–C bond formation) is rate-limiting (1, 9, 21). This assumption is also supported by KS–ACP cross-linking experiments, in which the extent of cross-linking (a direct measure of interaction between these two domains) correlates with chain elongation specificity (22). A representative example of the assay is shown in Fig. 2D with SHIV25, demonstrating the ability of the chimera to support higher levels of chain elongation activity with in combination with KS3 relative to KS6, with a strong preference for KS3. The chimeras grouped together by their [KS][AT] preference are shown in Fig. 3A (preference for [KS3][AT3] over [KS][AT6]) and Fig. 3B (vice versa). Analysis of the data revealed that substitution of the ACP6 derived HI (as in SHIV25), LII (as in AYC69), or HIII (as in AYC70) into an ACP3 sequence yielded a chimeric ACP with a similar [KS][AT] preference to ACP3 (Fig. 3A). Equivalently, substitution of the ACP3 derived HI (AYC67), LII (AYC73), or HIII (AYC74) into an ACP6 sequence yielded a chimeric ACP with a similar [KS][AT] preference to ACP6 (Fig. 3B). However, when the LI region was exchanged (as an HI-LI fragment in AYC68 or AYC72), the [KS][AT] preference of the resulting chimeric proteins was reversed (compare AYC68 with AYC67 and AYC72 with SHIV25). To directly test the influence of loop LI on chain elongation, two additional chimeras, SHIV22 and AYC79, were constructed in which only LI was exchanged between ACP6 and ACP3 (Fig. 3C and 3D). Both chimeras displayed changes in the preference for the [KS][AT] didomain that reflected the origin of LI.
Optimization of LI Boundaries: Defining the Minimal Epitope for Chain Elongation.
Although these results established that LI is the principal determinant of intramodular KS–ACP recognition, chimeric ACP proteins in which this loop region was exchanged (AYC68, AYC72, SHIV22, and AYC79) exhibited poorer activity relative to both wild-type ACP domains or even chimeras that harbored a contiguous LI–HII fragment. To identify possible causes for this deficiency, homology models of ACP3 and ACP6 were generated (23, 24) based on their high sequence identity (48–49%) to structurally characterized ACP2 (19). Examination of these models identified a triad of hydrophobic residues at the LI–HII junction (F47, L50 and L52 in ACP3, and F47, L50 and F52 in ACP6) that adopted significantly different conformations in the two proteins notwithstanding their sequence similarity. These residues were predicted to pack against each other in the core of the ACP near the Ser residue that serves as the attachment site of the phosphopantetheine arm (Fig. S3). We therefore hypothesized that a structurally perturbed LI–HII junction could account for the observed attenuation in activity of chimeras with exchanged LI regions. Consistent with this hypothesis, SHIV24, an AYC79-derived chimera in which the fusion junction was moved into the first turn of HII, showed increased activity with [KS6][AT6] (Fig. 3E)*. Importantly, SHIV29, the complementary chimera of SHIV24, was even more similar to wild-type ACP3 in its preference for KS3 over KS6 than was SHIV22 (Fig. 3F).
A final chimera, SHIV20 was constructed and analyzed to define precisely the relevant region of LI that determines ACP recognition during chain elongation. In this recombinant ACP domain, the HI–LI junction was shifted into LI (Fig. S4). SHIV20, which lacked the first five residues of LI from ACP3, was comparable to SHIV22 harboring full length LI. This suggests that the first few residues of LI play a marginal role, if any, in controlling chain elongation.
In summary, through a series of experiments summarized in Fig. 3, we were able to map to loop I of the ACP domain the principal determinants of KS–ACP recognition during polyketide chain elongation.
Molecular Basis for KS–ACP Recognition in Chain Elongation.
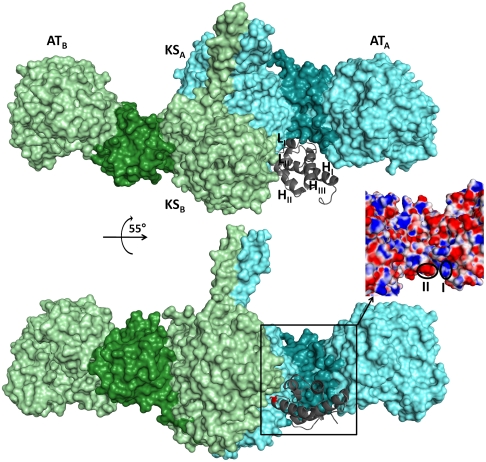
To gain further insight into the molecular basis for this striking molecular recognition, we have used the automated PatchDock (26, 27) and FireDock programs (28, 29) that utilize a combination of rigid body shape complementarity as well as modest side-chain reorientation for protein docking, to develop a model for how ACP5 interacts with the structurally characterized [KS5][AT5] didomain during polyketide chain elongation (for details, see Materials and Methods). The resulting model, shown in Fig. 4, accounts for all experimental data on mutual ACP–KS recognition reported both here and earlier (9). According to this model, the ACP domain from monomer A of the homodimer (ACPA) is docked in a deep cleft defined by the KSA domain, the KSA–ATA linker, and the ATA domain. Although ACPA participates in polyketide chain elongation in collaboration with KSB (30), both HII and most of LI of ACPA are tilted away from the KSB–ACPA interface. Instead, ACPA interacts with the parent [KS]A[AT]A subunit at two principal sites on the corresponding KSA–ATA linker, referred to as regions I and II (Fig. 4 and Fig. S5). Furthermore, the model predicts that regions I and II of the KSA–ATA linker interact with residues 44 and 45, respectively, in the LI loop of the ACPA domain. To test this model we constructed the R1420A/R1421A double mutant of DEBS module 3 (corresponding to R44A/R45A in the stand-alone ACP3 domain). As expected, the mutant showed a marked reduction in its ability to support chain elongation relative to wild-type module 3 (a 90% reduction in chain elongation activity; Fig. S6).
Fig. 4.
Docking model for ACP5 domain with the homodimeric [KS5][AT5] protein. Although the ACP domain of monomer A (gray) occupies a deep cleft between the KS and AT domains of monomer A (light cyan) (A), the conserved serine (red sticks) (B) at the N-terminal end of HII is positioned to participate in polyketide chain elongation with the KS active site of monomer B (light green). The KS-AT linker region of each monomer, which interacts with LI of the ACP domain (gray), is highlighted (dark cyan or dark green). The inset shows the two regions that show electrostatic complementarity with residue 44 (region I) and residue 45 (region II).
Examination of electrostatic potential maps for the corresponding regions of DEBS modules 3, 4 and 6 also revealed electrostatic complementarity in all cases between regions I and II and residues 44 and 45 in loop I of the corresponding ACP domain (Fig. 4 and Fig. S5). Importantly, these docking models also revealed a conserved hydrophobic patch near the entrance of the KS active site that could interact (< 5 Å) with a hydrophobic residue on helix II of the ACP located one turn downstream from the active site phosphopantetheinyl-bearing serine (Fig. S7). This observation too is consistent with our experimental data, summarized above.
The above model for interaction of the ACP domain with a core PKS module also provides a rationale for the universally conserved homodimeric architecture of a PKS module. Specifically, it predicts that catalysis of C–C bond formation is decoupled from recognition and proper matching of the correct KS and ACP partners that participate in this catalytic event. Whereas KS-catalyzed polyketide chain elongation requires access to the buried KS active site by the ACP-bound methylmalonyl moiety belonging to the paired subunit of the homodimer, proper KS–ACP recognition is dictated by interactions between the ACP domain and two specific regions located on the structured KS–AT linker within the same polyketide synthase subunit. Although KS domains are dimeric in both Type I and Type II PKSs, in the latter the KS exists as a discrete protein without a covalently attached KS–AT linker or AT domain. This suggests that the mode of KS–ACP recognition is different in these two systems and adds to a growing body of evidence suggesting important mechanistic differences between type I and II systems (31).
ACP Epitopes for Polyketide Chain Elongation and Transfer Are Distinct.
Having clearly defined the ACP epitope that controls polyketide chain elongation, we also wished to determine whether this same region also plays a role in controlling intermodular chain transfer, a distinct reaction in the overall catalytic cycle that involves correct pairing of ACPn and downstream KSn+1 partners. Previous studies examining chain transfer between DEBS modules 2 and 3, or alternatively between modules 4 and 5, revealed orthogonal KS-ACP specificity at these two intermodular junctions (8). We therefore asked whether intermodular chain transfer is also controlled by the same ACP epitope identified above that is responsible for the correct intramodular recognition in KS-catalyzed polyketide chain elongation.
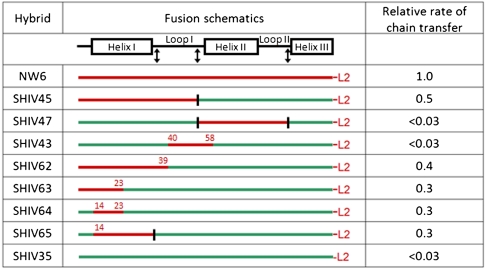
In earlier work, we developed a kinetic assay for the back-transfer of a diketide chain from a KS domain to an ACP domain (Fig. S2B), representing the microscopic reverse of the normal intermodular vectorial chain transfer reaction (7). We have now used this assay to measure the rate of back chain transfer from [KS3][AT3] to selected chimeras derived from ACP2 and ACP4 (Fig. 5). Surprisingly, the protein-protein interactions that control intermodular chain transfer could be localized to the first ten residues of the N-terminal helix (helix I) of ACP2 (compare SHIV64 to NW6)†. Thus, even though intermodular chain transfer and intramodular chain elongation both involve partnerships between the same KS active site and the electrophilic or nucleophilic pantetheinyl-bound substrate attached to two distinct but homologous ACP domains, the specific domain-domain interfaces that control these distinct biochemical reactions in the overall catalytic cycle are completely different (Fig. 6). It must be emphasized here that this difference is not simply manifest in the individual Michaelis constants (KM) of the two reactions, but is also reflected in the first-order rate constants (kcat) for the reactions between alternative KS and ACP pairs (7, 8). The detailed mechanism by which selective protein–protein interactions at domain surfaces influence the magnitude of transition state barriers for reactions occurring deep within the KS active site remains unknown.
Fig. 5.
Evaluation of chimeric ACP proteins in an assay for intermodular transfer of a growing polyketide chain. Chimeras of ACP2 and ACP4 were tested for their ability to accept a growing polyketide chain from KS3, the microscopic reverse of the normal intermodular chain transfer event, which is specific for ACP2 (8). The color scheme is: red = ACP2 derived, green = ACP4 derived. The rate of chain transfer for each chimera is normalized to corresponding value for the wild-type ACP2. “L2” refers to the C-terminal sequence of ACP2 that docks onto an N-terminal coiled-coil on module 3, and is essential for efficient intermodular chain transfer (35). The junctions utilized for constructing chimeras of ACP2 and ACP4 are defined in Fig. S8. Fusion sites distinct from the ones indicated by black arrows in Fig. S8 are color coded, and are defined by residue number consistent with the numbering in Fig. S8.
Fig. 6.
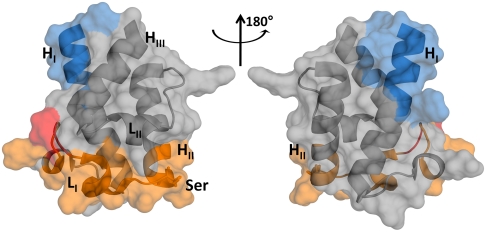
Distinct interfaces mediate protein–protein interactions. Chain elongation (orange with residues 44 and 45 shown in red) and chain transfer (blue) epitopes lie on entirely different faces of the ACP domain.
Conclusions and Summary.
In summary, combined experimental and computational analysis of selected DEBS modules demonstrates that pairwise KS–ACP recognition during intramodular polyketide chain elongation and intermodular polyketide chain transfer is controlled by distinct molecular recognition features, each of which appears to be highly conserved within the wider family of multimodular PKSs. These findings also underscore the occurrence of large-scale dynamic changes in these enzymatic assembly lines (18, 32–35). They also reveal an entirely unique principle in the modular control of polyketide antibiotic biosynthesis that is conceptually analogous but topologically unrelated to the protein-protein docking interactions that dictate DEBS1–DEBS2 and DEBS2–DEBS3 specificity (36). Specifically, our results indicate that not only are PKSs modular in that they are made up of domains of different structure and function, but the domains themselves have an intrinsically modular architecture in that they are made up of subdomains that have distinct roles in mediating interdomain interactions. This unique insight can be utilized to rationally engineer kinetically competent hybrid PKSs. Last but not least, we have also uncovered a clear functional rationale for the homodimeric architecture of multimodular PKSs. Parenthetically, we note that the reaction whose specificity depends on the dimeric architecture, C–C bond formation, lacks a counterpart in nonribosomal synthetases (NRPSs). In the latter family of assembly line proteins, peptide chain elongation and peptide transfer occur across an intermodular junction by a reaction that is equivalent to the intermodular chain transfer process in a PKS. Perhaps this explains why PKSs have evolved as homodimeric assemblies whereas the related NRPSs are monomers (37).
Materials and Methods
Cloning, Expression, and Purification of Constructs.
All constructs were cloned, expressed and purified using standard techniques or those published previously. A description of the procedures used is included in the SI Appendix. Oligonucleotide sequences used for the construction of chimeric ACPs and ACP mutants are included in Tables S1–S3 and Table S4, respectively. Circular dichroism spectra of a representative set of ACPs utilized in the chain elongation assay and the chain transfer assay are shown in Fig. S9 and Fig. S10, respectively.
Triketide Lactone Formation Assay to Determine [KS][AT] Activity in Chain Elongation.
See Fig. S2A. Each time point was set up in a reaction volume of 10 μL. [KS3][AT3] or [KS6][AT6] (10 μM, in 100 mM phosphate at pH 7.2) was incubated with 5 mM (2S,3R)-2-Methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester (NDK-SNAC) and 5 mM TCEP for 1 h at room temperature to acylate the KS domain. To measure the rate of chain elongation, 200 μM holo-ACP (wild-type or fusion protein) and 200 μM DL-[2-methyl-14C]-methylmalonyl-CoA were then added and allowed to react at room temperature. At each time point (either 30 or 40 min) the reaction was quenched by adding 10 μL of 1 M potassium hydroxide and heating the mixture for 20 min at 65 °C. Hydrochloric acid (10 μL of 1.5 M) was then added, and the mixture was dried in a Speed-vac for 2 h. The pellet was resuspended in 10 μL of ethyl acetate and spotted onto a TLC plate. A 60∶40 mixture of ethyl acetate/hexane was used for TLC, and the radiolabeled products were then visualized and quantified using a phosphorimager (Packard InstantImager).
Chain Transfer Assay to Determine Activity in Intermodular Chain Transfer.
See Fig. S2B. [KS3][AT3] (40 μM) was incubated with [1-14C]-NDK-SNAC (1.3 mM) and TCEP (5 mM) for 1 h at 22 °C in buffer A (100 mM phosphate, pH 7.2). Subsequent kinetic measurements were performed on ice and all components were prechilled. To measure the rate of chain transfer holo-ACP (wild-type or fusion protein) was then added to [14C] NDK labeled (3) [KS3][AT3] such that the final concentration of each protein was 25 μM in buffer B (100 mM phosphate, pH 7.2, 10% v/v glycerol). At each time point (30–150 s) samples were quenched with SDS-PAGE loading buffer lacking any reducing agents. The proteins were resolved on a 4–20% SDS-PAGE gradient gel (BioRad) and visualized with Coomassie blue stain. The gel was dried using a BioRad gel-drying system and analyzed using a phosphorimager (Packard InstantImager).
Molecular Docking Simulations and Electrostatic Potential Maps.
Docking simulations were carried out by using the PatchDock server (26, 27). The dimeric [KS5][AT5] didomain (PDB ID code: 2HG4) was defined as the receptor and the apo-ACP5 protein (homology model) was defined as the ligand. [KS5][AT5] didomain was used because it is the most complete high resolution structure from a type I PKS fragment solved to date (20). In contrast the [KS3][AT3] didomain crystal structure (38) lacks the N-terminal docking domain and has an irreversible inhibitor bound at the active site of the KS domain. The apo form of the ACP was used because it has been shown that for type I systems apo-, holo-, and acyl-ACPs are conformationally similar (31, 39). An additional distance constraint was applied such that the conserved serine on the ACP domain was between 12–25 Å of the active site cysteine of the KS domain to satisfy the constraint imposed by the length of the phosphopantetheine arm and search the KS surface more efficiently. The top 100 docking models identified by PatchDock were manually screened by utilizing the experimentally determined definition of the chain elongation epitope and 10 models were thus selected. All models had a remarkably similar ACP docking orientation as described in the text above. These 10 models were submitted to the FireDock server (28, 29) for structure refinement with the following parameters: Refinement type: full, atomic radius scale: 0.85, receptor and ligand residues were both unbound. Examination of the refined models consistently identified two sets of interacting sites between the loop I region of the ACP domain and the [KS][AT] didomain—residue 44 of ACPA with region I and residue 45 of ACPA with region II (Fig. 4 and Fig. S5, for [KS5][AT5] both regions lie on the KSA–ATA linker). A representative model (of 3 total that could be identified) that showed both sets of specific interactions is shown in Fig. 4.
Surface electrostatic potential maps were generated by using the APBS program (40). PDB2PQR (41, 42)was utilized to generate the input files for the APBS program utilizing the PARSE force field, and PROPKA program (43) to assign protonation states at pH = 7.2. PyMOL (44) was utilized to visualize the electrostatic surface maps as well as all other protein structures. Homology models for ACP3, ACP5, ACP6, [KS4][AT4], and [KS6][AT6] were generated by using the I-TASSER server (23, 24).
Supplementary Material
Acknowledgments.
We thank Sanketha Kenthirapalan for purifying constructs SHIV62-65 and Satoshi Yuzawa for helpful discussions. This work was supported by National Institutes of Health Grants GM 87934 (C.K.) and GM 22172 (D.E.C.), as well as Stanford Graduate Fellowships to S.K. and A.Y.C.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014081107/-/DCSupplemental.
*It is perhaps noteworthy that the last residue of the first turn of HII has also been implicated in KS–ACP specificity in Type II PKSs (25).
†We were unable to obtain a soluble chimera in which HIII from ACP2 is swapped into an ACP4 scaffold and therefore cannot rule out its involvement.
References
- 1.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu Rev Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 2.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel SC, Muller R. Formation of novel secondary metabolites by bacterial multimodular assembly lines: Deviations from textbook biosynthetic logic. Curr Opin Chem Biol. 2005;9:447–458. doi: 10.1016/j.cbpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 4.McDaniel R, Welch M, Hutchinson CR. Genetic approaches to polyketide antibiotics. 1. Chem Rev. 2005;105:543–558. doi: 10.1021/cr0301189. [DOI] [PubMed] [Google Scholar]
- 5.Hill AM. The biosynthesis, molecular genetics, and enzymology of the polyketide-derived metabolites. Nat Prod Rep. 2006;23:256–320. doi: 10.1039/b301028g. [DOI] [PubMed] [Google Scholar]
- 6.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 7.Wu N, Tsuji SY, Cane DE, Khosla C. Assessing the balance between protein–protein interactions and enzyme–substrate interactions in the channeling of intermediates between polyketide synthase modules. J Am Chem Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 8.Wu N, Cane DE, Khosla C. Quantitative analysis of the relative contributions of donor acyl carrier proteins, acceptor ketosynthases, and linker regions to intermodular transfer of intermediates in hybrid polyketide synthases. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- 9.Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2006;128:3067–3074. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim CY, et al. Reconstituting modular activity from separated domains of 6-deoxyerythronolide B synthase. Biochemistry. 2004;43:13892–13898. doi: 10.1021/bi048418n. [DOI] [PubMed] [Google Scholar]
- 11.Weissman KJ, Muller R. Protein–protein interactions in multienzyme megasynthetases. ChemBioChem. 2008;9:826–848. doi: 10.1002/cbic.200700751. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YM, Marrakchi H, White SW, Rock CO. The application of computational methods to explore the diversity and structure of bacterial fatty acid synthase. J Lipid Res. 2003;44:1–10. doi: 10.1194/jlr.r200016-jlr200. [DOI] [PubMed] [Google Scholar]
- 13.Weissman KJ, Hong H, Popovic B, Meersman F. Evidence for a protein–protein interaction motif on an acyl carrier protein domain from a modular polyketide synthase. Chem Biol. 2006;13:625–636. doi: 10.1016/j.chembiol.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Parris KD, et al. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure. 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YM, et al. Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III. J Biol Chem. 2001;276:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YM, Wu B, Zheng J, Rock CO. Key residues responsible for acyl carrier protein and beta-ketoacyl-acyl carrier protein reductase (FabG) interaction. J Biol Chem. 2003;278:52935–52943. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- 17.Rafi S, et al. Structure of acyl carrier protein bound to FabI, the FASII enoyl reductase from Escherichia coli. J Biol Chem. 2006;281:39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 19.Alekseyev VY, Liu CW, Cane DE, Puglisi JD, Khosla C. Solution structure and proposed domain domain recognition interface of an acyl carrier protein domain from a modular polyketide synthase. Protein Sci. 2007;16:2093–2107. doi: 10.1110/ps.073011407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci USA. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong FT, Chen AY, Cane DE, Khosla C. Protein–protein recognition between acyltransferases and acyl carrier proteins in multimodular polyketide synthases. Biochemistry. 2010;49:95–102. doi: 10.1021/bi901826g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapur S, et al. Mechanism based protein crosslinking of domains from the 6-deoxyerythronolide B synthase. Bioorg Med Chem Lett. 2008;18:3034–3038. doi: 10.1016/j.bmcl.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y, Lee TS, Kobayashi S, Khosla C. Ketosynthases in the initiation and elongation modules of aromatic polyketide synthases have orthogonal acyl carrier protein specificity. Biochemistry. 2003;42:6588–6595. doi: 10.1021/bi0341962. [DOI] [PubMed] [Google Scholar]
- 26.Duhovny D, Nussinov R, Wolfson HJ. Proceedings of the Second International Workshop, WABI 2002 Rome, Italy, September 17–21, 2002. In: Guigó R, Gusfield D, editors. Lecture Notes in Computer Science. Vol. 2452. Berlin: Springer; 2002. pp. 185–200. [Google Scholar]
- 27.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrusier N, Nussinov R, Wolfson HJ. FireDock: Fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 29.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao CM, Pieper R, Cane DE, Khosla C. Evidence for two catalytically independent clusters of active sites in a functional modular polyketide synthase. Biochemistry. 1996;35:12363–12368. doi: 10.1021/bi9616312. [DOI] [PubMed] [Google Scholar]
- 31.Ploskon E, et al. A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J Biol Chem. 2008;283:518–528. doi: 10.1074/jbc.M703454200. [DOI] [PubMed] [Google Scholar]
- 32.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 33.Tanovic A, Samel SA, Essen L-O, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 34.Koglin A, et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–911. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frueh DP, et al. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gokhale RS, Tsuji SY, Cane DE, Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 37.Sieber SA, et al. Evidence for a monomeric structure of nonribosomal peptide synthetases. Chem Biol. 2002;9:997–1008. doi: 10.1016/s1074-5521(02)00214-4. [DOI] [PubMed] [Google Scholar]
- 38.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran L, Broadhurst RW, Tosin M, Cavalli A, Weissman KJ. Insights into protein–protein and enzyme–substrate interactions in modular polyketide synthases. Chem Biol. 2010;17:705–716. doi: 10.1016/j.chembiol.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolinsky TJ, et al. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 44.DeLano W. The PyMOL User’s Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.