Abstract
G protein-activated inwardly rectifying potassium (GIRK or Kir3) channels are directly gated by the βγ subunits of G proteins and contribute to inhibitory neurotransmitter signaling pathways. Paradoxically, volatile anesthetics such as halothane inhibit these channels. We find that neuronal Kir3 currents are highly sensitive to inhibition by halothane. Given that Kir3 currents result from increased Gβγ available to the channels, we asked whether reducing available Gβγ to the channel would adversely affect halothane inhibition. Remarkably, scavenging Gβγ using the C-terminal domain of β-adrenergic receptor kinase (cβARK) resulted in channel activation by halothane. Consistent with this effect, channel mutants that impair Gβγ activation were also activated by halothane. A single residue, phenylalanine 192, occupies the putative Gβγ gate of neuronal Kir3.2 channels. Mutation of Phe-192 at the gate to other residues rendered the channel non-responsive, either activated or inhibited by halothane. These data indicated that halothane predominantly interferes with Gβγ-mediated Kir3 currents, such as those functioning during inhibitory synaptic activity. Our report identifies the molecular correlate for anesthetic inhibition of Kir3 channels and highlights the significance of these effects in modulating neurotransmitter-mediated inhibitory signaling.
Keywords: G Protein-coupled Receptors (GPCR), G Proteins, Neurotransmitters, Potassium Channels, Signal Transduction, Anesthetic, Gating
Introduction
Ion channels that control the excitability of neuronal conduction pathways are important targets for halogenated volatile anesthetics (HVAs)4 (1, 2). HVAs such as halothane enhance the activity of inhibitory GABAA and glycine receptors and inhibit excitatory channels such as glutamate and nicotinic receptors (3). Several two-pore domain K (K2P) channels are activated by HVAs (4–6). G protein-activated inwardly rectifying potassium (GIRK or Kir3) channels are also involved in inhibitory neurotransmission (7). Although ethanol and chloroform activate Kir3 channels (8, 9), paradoxically HVAs inhibit Kir3 channels (10, 11). Therefore, Kir3 channels present a rare case where an HVA such as halothane impairs inhibitory signaling by an ion channel.
The exact mechanism for modulation of ion channels by volatile anesthetics is not fully clear (2). Two residues in the TM2 and TM3 of GABAA participate in regulating the effects of enflurane (12). In nicotinic receptors, sites of action for halothane on tyrosine residues near the channel pore were identified (13). The transmembrane domains in AMPA and NMDA receptors play critical roles in their anesthetics and ethanol sensitivity (14, 15). Several sites on various K2P channels have been identified that are involved in anesthetic sensitivity (5, 16).
Inhibitory neurotransmitters, exemplified by GABA, stimulate Gαi/o-coupled receptors (17, 18) and liberate Gβγ that directly activates Kir3 channels (19). Kir3 channels mediate slow inhibitory post-synaptic currents in central neurons, limiting neuronal excitability (7). Kir3-null mice show increased seizure susceptibility (20), hyperalgesia (21), and reduced neurotransmitter-, morphine-, and ethanol-mediated anti-nociception (22, 23). Gβγ gating is a unique feature of the Kir3 family (24). Although like all other inward rectifiers, Kir3 channels require the membrane phospholipid phosphatidylinositol 4,5-bisphosphate for their activity (25, 26), they have weak phosphatidylinositol 4,5-bisphosphate interactions (27). Previous studies have identified regions in the C terminus (28) and the transmembrane domain (11) that contribute to halothane sensitivity but did not identify specific molecular correlates for the effect of halothane. Given the critical role of Kir3 channels in neurotransmission, we set out to determine the mechanistic basis for their paradoxical inhibition by halothane.
We find that halothane inhibits native hippocampal Kir3 channels and Kir3 expressed in HEK cells or Xenopus oocytes. Halothane inhibited both receptor-activated and basal currents. Halothane inhibition of Kir3 channels required Gβγ activation of the channel because removing Gβγ changed the direction of the effect of halothane from inhibition to activation. Furthermore, point mutants that reduced channel interaction with Gβγ reversed halothane inhibition. Finally, a single residue at the putative Gβγ gate controlled the effects of halothane on the channel.
EXPERIMENTAL PROCEDURES
Expression of Recombinant Channels in Xenopus Oocytes and HEK Cells
All constructs including Kir channels, channel mutants, human muscarinic type 2 receptors (hM2), C-terminal domain of β-adrenergic receptor kinase (cβARK), Gβ1, and Gγ2 were subcloned into the pGEMHE plasmid vector (29) for use in oocytes as described previously (30) and pcDNA3.1 for transfection into HEK cells as needed. Point mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene), and chimeras were made using splicing by overlap as described (30). The sequences of all constructs were confirmed by automated DNA sequencing. For oocyte expression, all constructs were linearized with appropriate restriction enzymes, and cRNAs were transcribed in vitro using mMessage mMachine (Ambion). cRNA concentration was estimated from two successive dilutions, which were electrophoresed in parallel on formaldehyde gels and compared with known concentrations of RNA marker (Invitrogen). cRNA concentration was adjusted to 320 ng/μl and diluted to desired concentrations on the day of injections. Expression of each protein was accomplished by injection of the desired amount of cRNA into Xenopus oocytes. In all two-electrode voltage clamp experiments, oocytes were injected with 1–2 ng of channel RNA and 2 ng of hM2, Gβ1, Gγ2, or cβARK where needed. Oocytes were isolated and microinjected as described previously (30). All oocytes were maintained at 18 °C, and electrophysiological recordings were performed 1–3 days following injection. HEK293 cells (ATCC) were plated on polylysine-treated coverslips and transfected 1 day later with appropriate DNA constructs. All cDNA transfections into HEK cells were performed using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions, and pEGFP-N3 vector (Clontech) was co-transfected with all constructs to identify successfully transfected cells using GFP fluorescence.
Hippocampal Neuron Preparation
Hippocampal neurons were prepared from embryonic day 18 rats following closely previously established methods (31). The cells were plated onto 12-mm polylysine-treated coverslips; whole-cell patch clamp was performed on 11–14 days in vitro cells.
Whole-cell Patch Clamp
Coverslips with neurons or HEK cells were moved to a chamber mounted onto an inverted Nikon microscope. Neurons were selected based on pyramidal shape body with multiple tapering processes and a thick apical dendrite. Successfully transfected HEK cells used for whole-cell patch clamp recordings were identified using fluorescence from co-transfected GFP. Recordings were performed using conventional whole-cell patch clamp. Electrodes (3–5 megaohms) were filled with intracellular solution (in mm): 140 KCl, 5 NaCl, 5 EGTA, 10 HEPES, 3 MgATP, and 0.2 Na2GTP (pH 7.4). The cells were superfused with low potassium (LK1) bath solution (in mm): 140 NaCl, 4 KCl, 2 CaCl2, 2 MgCl2, 20 HEPES, and 10 glucose, pH 7.4. Neurons were constantly voltage-clamped at −80 mV and recorded using a GapFree protocol. HEK cells were held at 0 mV and recorded using a ramp protocol of 1 mV/ms from −100 to +50 mV applied once every 250 ms. Currents were recorded using a Multiclamp700B amplifier, digitized with a Digidata 1322B, sampled at 4 kHz, low pass-filtered at 1 kHz, and collected using pClamp9.2 (all from Molecular Devices). Series resistance and cell capacitance were automatically compensated and monitored at the beginning and end of each experiment. Potassium currents were monitored by switching to a high potassium solution (HK1) containing (in mm): 5 NaCl, 140 KCl, 2 CaCl2, 1 MgCl2, 20 HEPES, and 10 glucose. All drugs were dissolved in high potassium and applied using an automated closed pressurized perfusion system with glass syringe reservoirs and Teflon tubing and valves. Current amplitudes were measured at −80 mV. Tertiapin-Q (120 nm), a specific peptide inhibitor of Kir3 channels, and barium (3 mm) were used to measure the residual inwardly rectifying current.
Two-electrode Voltage Clamp Recordings
Whole-oocyte currents were measured by conventional two-microelectrode voltage clamp with a GeneClamp 500 amplifier (Molecular Devices). Currents were recorded using a sampling rate of 1 kHz, low pass-filtered at 500 Hz, and collected using Clampex 9.0 (Molecular Devices). Data were analyzed using Clampfit 9.0 (Molecular Devices) and Origin 7.5 (OriginLab). Agarose-cushioned microelectrodes were used with resistances between 0.1 and 1.0 megaohms (32). Oocytes were constantly superfused with either a low potassium solution (LK2) having (in mm): 96 NaCl, 1 KCl, 1MgCl2, 5 HEPES (pH 7.4) or a high potassium solution (HK2) having (in mm): 96 KCl, 1 NaCl, 1MgCl2, 5 HEPES (pH 7.4). To block or activate currents, the oocyte chamber was perfused with solutions of the same composition with various drugs or 3 mm BaCl2 illustrated by bars above the tracings in the figures. Acetylcholine was used at 5 μm, and baclofen was used at 100 μm, both of which are saturating for stimulating their respective receptors. Typically, oocytes were held at 0 mV (EK), and a ramp protocol of 1 mV/ms from −100 to +100 mV was applied once every second.
Data and Statistical Analysis
Currents at −80 mV were measured for analysis and are presented in bar graphs and traces. All data are expressed as mean ± S.E. The effects of halothane are calculated as a percentage of the current in the absence of halothane in the same cell and expressed as the “percentage of halothane effect.” Dose responses were measured using single oocytes expressing each construct as noted in each figure. Dose-response curves were constructed using GraphPad Prism 5.0 (GraphPad) by constraining the minimum value at 0. Difference among groups was determined using t tests or one-way analysis of variance with Tukey's post hoc tests as appropriate and denoted in the figures. Differences of p < 0.05 were considered significant.
Anesthetic Preparation and Perfusion
Halothane was prepared as described elsewhere (33). Briefly, saturating concentrations were prepared by the addition of 0.2 g of halothane to 19 ml of recording solution in a scintillation vial filling most of the volume of the flask, minimizing the air gap inside the flask to prevent loss of anesthetic. This solution was vigorously shaken for 5 min and subsequently centrifuged at 800 × g at room temperature for 5 min. A fraction of the resulting saturated anesthetic solution was added to a volumetric flask to achieve the desired concentration of 0.5 mm, which is ∼2MAC (34). MAC is defined as the minimum alveolar concentration of inhaled anesthetic needed to suppress movement in 50% of subjects in response to a noxious stimulus (35). The flask was closed, and the solution was mixed by inverting 3–4 times. A closed pressurized perfusion system with glass syringe reservoirs and Teflon tubing and valves were used to apply all the solutions to the mammalian cells or oocytes. Fresh working solutions were used at the start of each recording.
RESULTS
Halothane Inhibits Native and Expressed Kir3 Channels
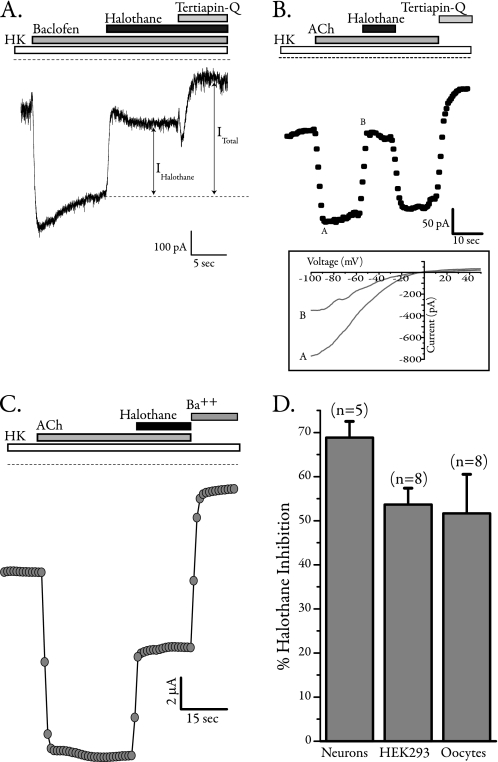
In hippocampal neurons, application of the GABAB agonist baclofen activates tertiapin-Q-sensitive Kir3 currents that are robustly inhibited by 0.5 mm (2MAC) halothane (Fig. 1A). Because hippocampal neurons most likely express a combination of Kir3.1 and Kir3.2, we used HEK293 cells and Xenopus oocytes to express these channels with a different G protein-coupled receptor, human muscarinic 2 (hM2), to assess the effects of halothane. In HEK cells and oocytes, 2MAC halothane inhibited acetylcholine-(ACh-) activated and tertiapin-Q- and Ba2+-sensitive Kir3.1/3.2 currents (Fig. 1, B and C). Summary data for halothane inhibition of Kir3.1/3.2 currents in native and heterologous expression systems are shown in Fig. 1D. Halothane strongly and to a similar extent inhibited currents mediated by two different G protein-coupled receptors in three cell systems.
FIGURE 1.
Halothane inhibits native and expressed Kir3 channels. A, sample whole-cell trace from 14 days in vitro hippocampal neuron recorded at −80 mV. Stimulation of GABAB receptors using baclofen activated a large inward current that was inhibited by halothane. The remaining current was blocked by tertiapin-Q, a selective peptide blocker of Kir3 channels. B, whole-cell current from a HEK cell expressing Kir3.1/3.2 with human muscarinic 2 (hM2) receptors. Currents were monitored using a voltage ramp (inset), and values at −80 mV were plotted. Activation of hM2 receptors by ACh resulted in large inwardly rectifying currents, which were robustly inhibited by co-application of halothane. All currents were subsequently blocked by tertiapin-Q. HK, high potassium. C, two-electrode voltage clamp recording of Kir3 currents from Xenopus oocytes expressing Kir3.1/3.2 and hM2. Cells were perfused with high potassium solution, receptors were stimulated using 5 μm ACh, and large inwardly rectifying currents at −80 mV are plotted that were robustly inhibited by co-application of halothane. D, summary data for inhibition of Kir3 current by halothane in various cell types (neurons, n = 5; HEK cells, n = 8; oocytes, n = 8).
Halothane Inhibition Requires Intact Gβγ Interactions with the Channel
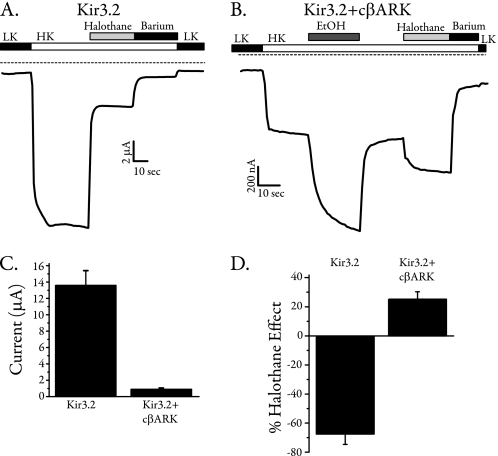
Because Kir3 channels are the only inward rectifiers that are G protein-sensitive, we asked whether the G protein-insensitive Kir channels that are highly homologous to Kir3 channels are halothane-sensitive. A previous report showed that Kir1.1 and Kir2.1 are fairly insensitive to the effect of halothane in oocytes (11). We expressed Kir2.1, Kir2.2, Kir2.3, Kir2.4, Kir4.1, and Kir7.1 in oocytes and determined their relative sensitivity to 0.5 mm halothane. Kir2.1 and Kir4.1 were slightly activated by halothane (∼8 and 3% activation for each), Kir2.4 was completely insensitive to halothane, and Kir2.2, Kir2.3, and Kir7.1 were moderately inhibited by halothane (∼5, 6, and 10% inhibition for each, respectively). Summary data for halothane sensitivity of these channels is shown in supplemental Fig. S1. Because Gβγ activation is a hallmark difference between Kir3 and other inward rectifiers, we asked whether Gβγ interaction with the channel may be a target for halothane. Using a homomeric channel would better facilitate direct analysis of molecular sites involved in halothane modulation. We therefore used Kir3.2 alone in these experiments. Kir3.2 is abundantly expressed in the brain as a highly functional homomeric channel (36–39). We expressed Kir3.2 channels in Xenopus oocytes with and without the membrane-targeted cβARK (aka cGRK2), which scavenges free Gβγ (40, 41). Consistent with previous results, cβARK expression significantly reduced the basal current (compare tracing in Fig. 2, A and B; summary shown in Fig. 2C) and essentially eliminated ACh-induced currents as reported (42), indicating effective scavenging of Gβγ. As expected, currents from oocytes expressing Kir3.2 alone were very robustly inhibited by halothane (Fig. 2A). In the presence of cβARK, ethanol (200 mm) activated robust currents, confirming that ethanol activated the channel independently of Gβγ (43). Surprisingly, application of halothane to these oocytes also increased channel activity. Summary data for the effects of halothane on Kir3.2 in the presence or absence of cβARK are shown in Fig. 2D. Co-expression of cβARK also reversed halothane inhibition in oocytes expressing Kir3.1/3.2 (supplemental Fig. S2). Therefore, expression of cβARK (i.e. scavenging Gβγ) reversed the effect of halothane from inhibition to activation, strongly suggesting a role for Gβγ activation as a target for halothane-induced inhibition.
FIGURE 2.
Scavenging Gβγ reverses halothane inhibition of Kir channels. A, two-electrode voltage clamp recordings from an oocyte expressing Kir3.2 alone, where halothane robustly inhibits channel activity. LK, low potassium; HK, high potassium. B, two-electrode voltage clamp recordings from an oocyte expressing Kir3.2 and the membrane-targeted pleckstrin homology domain of cβARK, which strongly binds and scavenges Gβγ. Currents were significantly smaller in the presence of cβARK (note different scale from A), indicating the effectiveness of cβARK. In these oocytes, both ethanol and halothane activated the channel. C, summary data showing the effectiveness of cβARK co-expression in reducing Kir3.2 currents (control, n = 5; cβARK co-expressed, n = 11). D, summary data comparing the effects of halothane on Kir3.2 alone and Kir3.2 co-expressed with cβARK. Scavenging Gβγ reversed the effects of halothane (control, n = 5; cβARK co-expressed, n = 11).
Mutants That Impair Gβγ Interaction with the Channel Reverse the Effects of Halothane
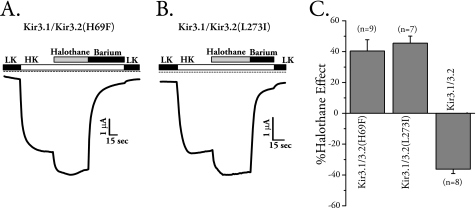
To further scrutinize the dependence of halothane-mediated inhibition on Gβγ interactions, we mutated two sites on Kir3 channels that we have shown in the past to be critical in both binding to and activation by Gβγ (44). Mutation of Leu-268 to Ile or His-64 to Phe in isolated C or N terminus of Kir3.4 impairs Gβγ binding and activation (44). Another study showed that corresponding mutants in Kir3.2 (H69F and L273I) also impair Gβγ activation of the channel, whereas these authors did not observe a change in G protein binding (45). We made these mutations and tested the effects of halothane. Expression of Kir3.2 homomers with these mutations had very small currents that could not be reliably measured, consistent with findings in Kir3.4 channels (44). We therefore expressed wild-type Kir3.1 with each mutant and tested the sensitivity of channels containing Kir3.2(L273I) or Kir3.2(H69F) toward halothane. Channels with either mutant were activated by halothane similar to that seen for channels co-expressed with cβARK (Fig. 3, A–C). Taken together these results indicate that halothane inhibition of Kir3.1/3.2 required proper functional Kir3.2 channel-Gβγ interaction because disruption of this activation by scavengers such as cβARK or by point mutations on the channel reversed the effects of halothane.
FIGURE 3.
Mutants that impair Gβγ interaction with the channel reverse the effects of halothane. The Kir3.2 mutants H69F and L273I were made based on previously identified sites critical for Gβγ interaction in Kir3.1 and Kir3.4. Each mutant was co-expressed in oocytes with wild-type Kir3.1 because expression alone or co-expression with similarly mutated Kir3.1 resulted in very small currents that could not be reliably quantified. A, currents from Kir3.2(H69F) co-expressed with Kir3.1 were activated by halothane. LK, low potassium; HK, high potassium. B, currents from Kir3.2(L273I) co-expressed with Kir3.1 were activated by halothane. C, summary results for the effects of mutants that impaired Gβγ interaction on halothane sensitivity of Kir3.2. Both H69F and L273I reversed the halothane inhibition (n = 7–9).
Halothane Affects Gβγ-mediated Gating of Kir3 Channels
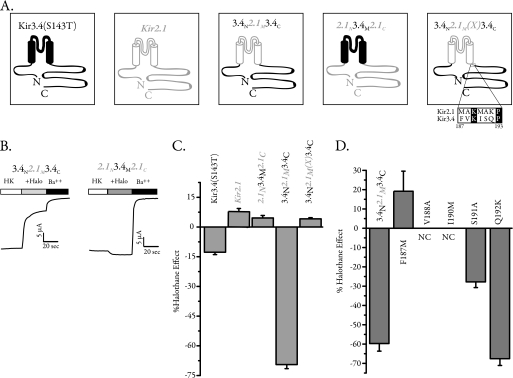
A previous study (11) used several chimeras between Kir2.1 and Kir3.2 in an attempt to identify halothane-sensitive elements in Kir3.2 channels. This study found a large region (residues 87–223), which encompasses the entire membrane domain, to be involved in halothane sensitivity. Small replacements in the pore and TM2 region showed that the pore and the first half of the TM2 are not involved in halothane sensitivity. Another study showed that deletion of a large portion of the Kir3.2 C terminus removed the effect of halothane (28). Biochemically, they showed that volatile anesthetics did not change Gβγ binding to the channel C terminus in GST pulldown assays. However, neither study identified the exact molecular correlate for the effects of halothane. Kir2.1 is Gβγ-insensitive and moderately activated by halothane, whereas Kir3.4 is Gβγ-sensitive and inhibited by halothane, albeit less than Kir3.2 (supplemental Fig. S1). We therefore speculated that a chimeric construct between Kir2.1 and Kir3.4 would flesh out the regions involved in halothane modulation. We used Kir3.4(S143T) that harbors a point mutation in the pore region. This mutant shows robust homomeric channel activity and has proven a powerful tool to study structure-function relationship in several studies (42, 46). We have previously used chimeras between Kir3.4(S143T) and Kir2.1 channels to identify G protein and phosphatidylinositol 4,5-bisphosphate-interacting regions on these channels (25, 42). Two of these chimeras, 3.4N2.1M3.4C and 2.1N3.4M2.1C, swap the transmembrane and cytosolic regions of Kir2.1 and Kir3.4(S143T). 3.4N2.1M3.4C harbors the N and C termini of Kir3.4 and the membrane region of Kir2.1,Y, and 2.1N3.4M2.1C is the reverse chimera (Fig. 4A). Both chimeras showed strong inwardly rectifying currents, and although 2.1N3.4M2.1C was essentially insensitive to the effect of halothane, 3.4N2.1M3.4C was robustly inhibited by halothane (Fig. 4B). Summary data for halothane effects on the two chimeras and their parent constructs are shown in Fig. 4C. This strongly suggested that the halothane inhibition arises from the cytosolic regions. However, the chimera 3.4N2.1M3.4C contained amino acid residues 186–419 from Kir3.4, and recently available structures suggest that the cytosolic region of the channel starts several amino acids after this junction (47–49). We therefore made a new construct, 3.4N2.1M(X)3.4C, which moved the chimera junction to include an extra 6 amino acids from Kir2.1 (Fig. 4A). We expressed this chimera in oocytes and found that the halothane inhibition was completely abolished (Fig. 4C). An alignment of the additional region added in 3.4N2.1M(X)3.4C shows that 5 amino acids are different between the two channels in this region (Fig. 4A); we mutated each amino acid, one at a time, in 3.4N2.1M3.4C to the corresponding Kir2.1 residue and tested for halothane sensitivity. Two of the mutants failed to produce significant current, and although S191A and Q192K were both inhibited by halothane, the mutant F187M completely abolished halothane inhibition and was in fact activated by halothane (Fig. 4D).
FIGURE 4.
Chimeric approach to identify the determinant for halothane sensitivity in Kir3 channels. Kir2 channels are very modestly activated by halothane, whereas Kir3 channels are inhibited. We used chimeras between Kir2.1 and Kir3.4 to swap the membrane and intracellular domains to identify exact molecular determinants for halothane sensitivity. A, schematic depiction of the two parent channels, the monomeric active Kir3.4(S143T) and Kir2.1, as well the two chimeras in which intracellular domains were swapped between the two channels. The two-transmembrane domains are depicted as cylinders, and channel pieces are color-coded in open gray (Kir2.1) or solid black (Kir3.4). For the last chimera (3.4N2.1M(X)3.4C), the region at the end of the Kir2.1M in 3.4N2.1M3.4C was extended to include 6 additional amino acids from Kir2.1. B, sample TEVC recording from two oocytes expressing 3.4N2.1M3.4C and 2.1N3.4M2.1C and recording the effects of halothane application (+Halo) on each one. Although 2.1N3.4M2.1C was essentially insensitive to halothane, 3.4N2.1M3.4C currents were robustly inhibited by halothane. HK, high potassium. C, summary data for halothane effect on Kir3.4(S143T), Kir2.1, Kir3.4N2.1M3.4C, Kir2.1N3.4M2.1C, and Kir3.4N2.1M(X)3.4C. The extended region of Kir2.1 completely abolished the halothane inhibition. D, mutation of each of the amino acids in the extended region from Kir3.4 residue to that of Kir2.1 in the 3.4N2.1M3.4C chimera (panel A). Although two mutants did not produce appreciable currents and S191A and Q192K were inhibited by halothane, F187M not only completely removed halothane inhibition but was moderately activated. NC, no current.
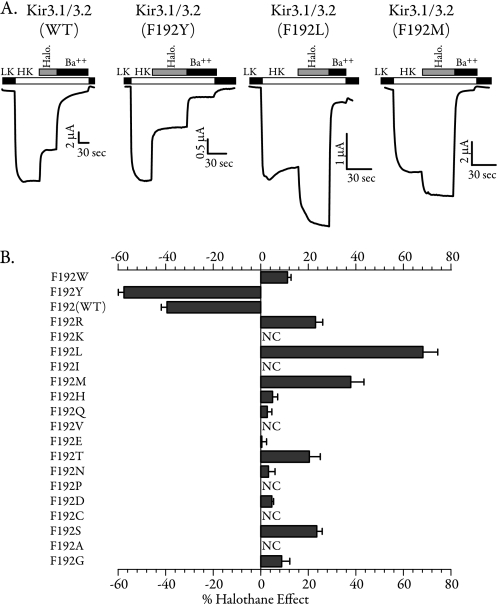
Mutating the Residue at the Putative Gate of Kir3.2 Reverses Halothane Inhibition
Phenylalanine 187 in this chimera corresponds to Phe-187 in Kir3.4, which we previously identified as the putative Gβγ-controlled gate of the channel (50). In Kir3.2, which is the major neuronal subtype and most robustly inhibited by halothane, Phe-192 occupies this putative gate position. We therefore asked whether the Phe-192 residue in Kir3.2 also controls halothane sensitivity. Mutation of Phe-192 to a methionine (the residue in Kir2.1) resulted in a reversal of halothane inhibition where Kir3.2(F192M) mutant was robustly activated by halothane (Fig. 5A).
FIGURE 5.
Halothane activates Kir3.2(F192M) and Kir3.2(L273I) in a dose-responsive manner. A, sample TEVC recording from an oocyte expressing Kir3.1/Kir3.2(F192M) and its response to varying concentrations of halothane. The oocyte was held at −80 mV, and currents were measured in high potassium (HK). Varying concentrations of halothane were applied, and barium was used at the end to determine basal currents. The dose-response curve for halothane activation of this channel is seen on the right. The EC50 for activation was 0.151 mm. B, sample TEVC recording from an oocyte expressing Kir3.1/Kir3.2(L273I) and its response to varying concentrations of halothane. The EC50 for activation was 0.862 mm.
Halothane Activates Kir3.2(F192M) and Kir3.2(L273I) in a Dose-responsive Manner
A previous report (28) showed that Kir3.1(F137S), a homomeric active mutant channel (46), is activated by high concentrations of halothane (1 mm), whereas it is inhibited by low concentrations of halothane (100 μm). In light of these findings, we performed halothane dose-response curves for channels harboring the L273I or F192M mutation. Halothane activated both channels at all doses tested in a concentration-dependent manner. Traces and summary dose-response curves for each one are shown in Fig. 5.
Kir3.2 Gate Mutant Is Gβγ-sensitive
Because changing Gβγ interactions altered channel modulation by halothane, we also asked whether the gate mutant F192M is sensitive to Gβγ activation. To that end, we expressed Kir3.1/3.2 or Kir3.1/3.2(F192M) with or without exogenous Gβ1γ2 and found that indeed the F192M mutant was Gβγ-sensitive (Fig. 6A). Because this mutant was activated by both Gβγ and halothane, we asked whether the presence of Gβγ co-expression or receptor activation would alter halothane sensitivity. Using Kir3.1/3.2(F192M) expressed with hM2 or Gβ1γ2, we measured halothane sensitivity. Halothane activated basal currents, ACh-induced currents, and currents in the presence of exogenous Gβ1γ2. However, both ACh-induced and Gβγ-enhanced currents were significantly less activated by halothane (Fig. 6, B and C).
FIGURE 6.
The gate mutant F192M is Gβγ-sensitive and activated by both receptor stimulation and halothane. A, bar graph summary from TEVC recordings in oocytes expressing Kir3.1/3.2 or Kir3.1/3.2(F192M) with or without exogenously expressed Gβ1γ2. Kir3.1/3.2(F192M) is sensitive to G proteins because basal currents for this mutant are increased in the presence of co-expressed Gβγ similar to those seen in wild-type channels. B, sample TEVC recording from an oocyte expressing Kir3.1/Kir3.2(F192M)/hM2 and its response to ACh and halothane. A stacked bar graph on the right shows average data from eight similar recordings. LK, low potassium; HK, high potassium. C, summary data for halothane activation of Kir3.1/3.2(F192M) under basal, receptor-stimulated, and Gβγ-activated conditions. Response after receptor stimulation or in the presence of exogenous Gβγ was reduced (* denotes p < 0.05, one-way analysis of variance, with Tukey's post hoc test).
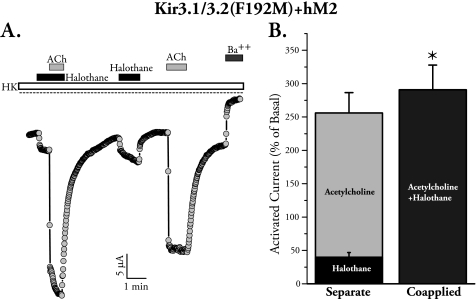
Halothane and Gβγ Show Synergy in Activating Kir3.2(F192M)
We then asked whether the effects of Gβγ and halothane are additive, occlusive, or synergistic in the F192M mutant. We expressed Kir3.1/3.2(F192M) with hM2 and measured halothane- and ACh-induced currents when applied separately and when applied together in the same oocyte. Co-application of halothane and ACh, regardless of order of application, resulted in a modest but significantly more than additive response, indicating some synergy between the two activators (Fig. 7A). Summary data are shown in Fig. 7B. A sample tracing where application order was reversed is shown in supplemental Fig. S3.
FIGURE 7.
Activation of the gate mutant F192M by receptor stimulation and halothane is more than additive. A, sample TEVC recording from an oocyte expressing Kir3.1/Kir3.2(F192M)/hM2 and its response to ACh and halothane applied together or separately. HK, high potassium. B, bar graph summary for current induction by co-application of halothane and ACh as compared with the combined effect of the two applied separately. The value for the two co-applied is significantly larger that than the effects of the two added separately (* denotes p < 0.05, paired t test, n = 8).
We also tested the effects of halothane on Kir3.2(S177T), which has constitutive activity (51). This mutant behaves similarly to those we had identified in Kir3.4 (50) whose basal currents were Gβγ-independent. Halothane activated Kir3.2(S177T), and summary data are shown in supplemental Fig. S4.
Identity of the Residue at the Putative Channel Gate Alters Its Response to Halothane
Because the mutant F192M reversed the halothane inhibitory effect, we tested whether other substitution at this site would have a similar effect. We mutated Phe-192 to each of the other 18 amino acids and tested their sensitivity to halothane in the presence of Kir3.1. Sample traces from the wild-type channel and three of the mutants are shown in Fig. 8A. Summary data for all mutants are shown in Fig. 8B. A number of Kir3.2 mutants did not produce significant currents when co-expressed with Kir3.1 (NC, no current). The presence of phenylalanine (wild type) or tyrosine at this position resulted in channel inhibition by halothane. All other mutants were either activated or not affected by halothane. We tested the effects of enflurane (supplemental Fig. S5) and isoflurane (not shown) on three of the gate mutants. We found that these two anesthetics inhibited F192Y and stimulated F192M and F192L mutants, similar to the halothane results. This indicates that the findings on halothane can be generalized to other volatile anesthetics.
FIGURE 8.
Halothane affects Gβγ-mediated gating of Kir3 channels. A, TEVC recordings from oocytes expressing Kir3.1/3.2 as well as three of the mutants tested. Although halothane robustly inhibited the wild type and F192Y, it activated mutants F192L and F192M. LK, low potassium; HK, high potassium. B, the bar data show the summary of halothane sensitivity for all 20 amino acids at position 192. All the mutants were tested with wild-type Kir3.1 (n = 8–12). The residues are listed in the order of amino acid volume (largest to smallest). NC denotes no current.
DISCUSSION
We have identified Gβγ-mediated activation as the underlying target for Kir3 channel inhibition by halothane. Scavenging Gβγ away from the channel or mutating Gβγ-interacting residues on Kir3.2 not only completely abolished inhibition but caused channel activation by halothane. The specific effect on Gβγ-mediated gating and identification of residues on Kir3 channels whose mutation fully reverses the effects of halothane are one of the most compelling findings for specific molecular interactions of an anesthetic with a physiologically relevant target. In addition, we can now address the mechanism by which anesthetics exert their effects on these channels.
Our data support a model where halothane acts through Phe-192 as a partial agonist (i.e. weak gating molecule), whereas Gβγ is a full agonist (strong gating molecule). Therefore, in the presence of adequate Gβγ activation, the addition of halothane inhibits channel activity, similar to the action of a partial agonist in the presence of a full agonist. When Gβγ activation is reduced, through mutations or by co-expression of βARK, halothane activates the channels as a partial agonist. In constitutively active channels such as S177T, the basal activity is mainly Gβγ-insensitive, so the channels can be activated by halothane. A methionine residue at position 192 increases the efficacy of halothane as an agonist and reveals channel activation under basal conditions. Under these conditions, excess Gβγ reduces the effects of halothane. Finally, the difference in sensitivity that is observed for halothane among Kir3 channel subunits could be due to their differential affinity to both halothane and Gβγ. Previous studies have shown that distinct Kir3 subunits contribute differentially to the Gβγ activation of the channels, and this may be due to different Gβγ affinities of each channel subunit (44, 45, 52, 53). Therefore, in Kir3.2 channels, Gβγ activation may be stronger, and halothane is a less efficacious partial agonist, leading to the largest inhibition. A previous study (28) found that halothane has different effects on Kir3.1 at low and high concentrations. Our model explains those findings where the activation of halothane as a partial agonist would only be manifested at high concentrations because Gβγ activation of homomeric Kir3.1 may not be as efficacious as those for other subtypes, especially under basal conditions tested in that study (28).
Channels activated by receptor-stimulated Gβγ were most robustly inhibited by halothane. Neurotransmitter stimulation of G protein-coupled receptors in neurons is critically important in shaping synaptic activity. Although some basal Kir3 channel activity in dendrites has been reported (54), the majority of Kir3 current is activated only after neurotransmitter stimulation (55, 56). That this neurotransmitter-mediated current was robustly inhibited by halothane suggests that these channels may be important targets for in vivo anesthetic action. However, the paradoxical inhibition by halothane, which would have a net excitatory effect, implicates Kir3 currents as contributors to side effects of anesthesia (57). Alternatively, such effects could be taking place in inhibitory interneurons that may express Kir3 channels. A recent study showed that isoflurane causes activation of some chemosensory neurons through inhibition of a THIK-1-like K2P channel that may be essential in maintaining some respiratory motor control during anesthesia (58). This presents another example of seemingly paradoxical potassium channel inhibition by anesthetics, which may control a specific physiological response. In Kir3.2-null mice, pain threshold is lowered (23), and morphine-mediated antinociception is reduced (22). Interestingly, opioids, which activate Kir3 channels in neurons (59), are commonly co-administered with inhalation anesthetics during surgery; therefore, inhalation anesthetics and opioids could exert opposing effects on Kir3 channel activity.
GABAA and GABAB receptors constitute chloride-selective channels and Gi/o-coupled receptors, respectively. GABA release activates GABAA receptors, and this effect is directly stimulated by anesthetics, is fast, and is localized to the synapse, requiring large concentrations of GABA. GABAB receptors, on the other hand, are mostly extrasynaptic, have higher affinity for GABA, and are believed to be activated due to GABA “spillover” from the synapse (60). This GABA-mediated signal would proceed through effectors such as Kir3 channels to shape slow inhibitory signaling (39). Inhibition of this signaling pathway by anesthetics may serve to fine-tune the effectiveness of anesthetics or to reverse their effectiveness and manifest as untoward side effects.
Having a phenylalanine or a tyrosine at the channel gate led to channel inhibition by halothane. Several protein structures that have been solved in the presence of anesthetics suggest that they may prefer amphipathic environments (61). Additionally, aromatic residues have been shown to interact with halothane in photo-labeling studies (13). We speculate that the phenyl ring in the phenylalanine or tyrosine residues may engage in hydrophobic interactions with halothane, whereas the surrounding water or the hydroxyl group on tyrosine could hydrogen-bond with halothane. This site at the helix bundle crossing in the transmembrane domain of the channel comprises the Gβγ-operated gate (50); hence, halothane binding to the aromatic residue here could control gating. This amino acid residue is contained within the larger regions identified in previous studies that affected halothane sensitivity (11, 28). Consistent with those reports, our findings suggest that the agonist-mediated currents are most highly sensitive to the effects of halothane. Physiologically, neuronal Kir3 channels have very robust neurotransmitter-activated currents (55, 56). The sensitive nature of the agonist-mediated current implicates these channels as clinically important targets for anesthetic action.
The most compelling studies on the sites of action for anesthetics on ion channels point to regions at or close to the transmembrane domains (2). What is less clear is whether anesthetics exert specific effects on channel gating. Milovic et al. (28) speculated that halothane may interfere with the gating mechanism of the channel. Our finding that the Kir3.2 channel gate itself is a critical determinant of anesthetic action provides a novel mechanistic target for these clinically important molecules. Although Cys-loop channels, such as nicotinic, GABA, and glycine receptors, are not structurally very close to Kir channels, two other important anesthetic-sensitive channel families, namely ionotropic glutamate receptors and K2P channels, are. The pore of glutamate receptor resembles an upside-down Kir channel pore (62), and K2P channels resemble tandem repeats of Kir channels. For both classes of channels, residues in the transmembrane segments and their proximal regions have been identified that alter anesthetic sensitivity (5, 14, 16). The most compelling residue identified is at position 159 in two-pore domain acid-sensitive potassium (TASK) channels that is located at the start of the third transmembrane domain, which in these channels faces the cytoplasm (16). Residues at the end of transmembrane 4 in these channels have also been implicated in the action of anesthetics (5). It was reported that G proteins can directly modulate K2P channels (63), and although both K2P and Kir3 channels require phosphatidylinositol 4,5-bisphosphate for their activity, the effects of halothane may not necessarily depend on these interactions but rather on specific residues at or close to the channel gate itself. The exact mechanism of gating for K2P channels is not yet clear; however, it is tempting to speculate that the gate in these and perhaps other channels may regulate anesthetic action similar to what we find in Kir3 channels.
Supplementary Material
Acknowledgments
We thank Dr. E. Reuveny for cβARK and Dr. M. Lazdunski for Kir3.2. We thank Dr. Coeli Lopes for advice on anesthetic solution preparation and perfusion.
This work was supported, in whole or in part, by American Heart Association-Great Rivers Affiliate Beginning Grant-in-Aid 0765275U and by funds from the Geisinger Clinic (to T. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- HVA
- halogenated volatile anesthetic
- GABA
- γ-aminobutyric acid
- βARK
- β-adrenergic receptor kinase
- MAC
- minimum alveolar concentration
- ACh
- acetylcholine
- K2P
- two-pore domain K
- TEVC
- two-microelectrode voltage clamp.
REFERENCES
- 1.Campagna J. A., Miller K. W., Forman S. A. (2003) N. Engl. J. Med. 348, 2110–2124 [DOI] [PubMed] [Google Scholar]
- 2.Franks N. P. (2006) Br. J. Pharmacol. 147, Suppl. 1, S72–S81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemmings H. C., Jr, Akabas M. H., Goldstein P. A., Trudell J. R., Orser B. A., Harrison N. L. (2005) Trends Pharmacol. Sci. 26, 503–510 [DOI] [PubMed] [Google Scholar]
- 4.Patel A. J., Honoré E., Lesage F., Fink M., Romey G., Lazdunski M. (1999) Nat. Neurosci. 2, 422–426 [DOI] [PubMed] [Google Scholar]
- 5.Talley E. M., Bayliss D. A. (2002) J. Biol. Chem. 277, 17733–17742 [DOI] [PubMed] [Google Scholar]
- 6.Franks N. P., Honoré E. (2004) Trends Pharmacol. Sci. 25, 601–608 [DOI] [PubMed] [Google Scholar]
- 7.Lüscher C., Jan L. Y., Stoffel M., Malenka R. C., Nicoll R. A. (1997) Neuron 19, 687–695 [DOI] [PubMed] [Google Scholar]
- 8.Lewohl J. M., Wilson W. R., Mayfield R. D., Brozowski S. J., Morrisett R. A., Harris R. A. (1999) Nat. Neurosci. 2, 1084–1090 [DOI] [PubMed] [Google Scholar]
- 9.Aryal P., Dvir H., Choe S., Slesinger P. A. (2009) Nat. Neurosci. 12, 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigl L. G., Schreibmayer W. (2001) Mol. Pharmacol. 60, 282–289 [DOI] [PubMed] [Google Scholar]
- 11.Yamakura T., Lewohl J. M., Harris R. A. (2001) Anesthesiology 95, 144–153 [DOI] [PubMed] [Google Scholar]
- 12.Mihic S. J., Ye Q., Wick M. J., Koltchine V. V., Krasowski M. D., Finn S. E., Mascia M. P., Valenzuela C. F., Hanson K. K., Greenblatt E. P., Harris R. A., Harrison N. L. (1997) Nature 389, 385–389 [DOI] [PubMed] [Google Scholar]
- 13.Chiara D. C., Dangott L. J., Eckenhoff R. G., Cohen J. B. (2003) Biochemistry 42, 13457–13467 [DOI] [PubMed] [Google Scholar]
- 14.Minami K., Wick M. J., Stern-Bach Y., Dildy-Mayfield J. E., Brozowski S. J., Gonzales E. L., Trudell J. R., Harris R. A. (1998) J. Biol. Chem. 273, 8248–8255 [DOI] [PubMed] [Google Scholar]
- 15.Ronald K. M., Mirshahi T., Woodward J. J. (2001) J. Biol. Chem. 276, 44729–44735 [DOI] [PubMed] [Google Scholar]
- 16.Andres-Enguix I., Caley A., Yustos R., Schumacher M. A., Spanu P. D., Dickinson R., Maze M., Franks N. P. (2007) J. Biol. Chem. 282, 20977–20990 [DOI] [PubMed] [Google Scholar]
- 17.Sodickson D. L., Bean B. P. (1996) J. Neurosci. 16, 6374–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torrecilla M., Marker C. L., Cintora S. C., Stoffel M., Williams J. T., Wickman K. (2002) J. Neurosci. 22, 4328–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logothetis D. E., Kurachi Y., Galper J., Neer E. J., Clapham D. E. (1987) Nature 325, 321–326 [DOI] [PubMed] [Google Scholar]
- 20.Signorini S., Liao Y. J., Duncan S. A., Jan L. Y., Stoffel M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 923–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marker C. L., Cintora S. C., Roman M. I., Stoffel M., Wickman K. (2002) Neuroreport 13, 2509–2513 [DOI] [PubMed] [Google Scholar]
- 22.Mitrovic I., Margeta-Mitrovic M., Bader S., Stoffel M., Jan L. Y., Basbaum A. I. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blednov Y. A., Stoffel M., Alva H., Harris R. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirshahi T., Jin T., Logothetis D. E. (2003) Sci. STKE 2003, PE32. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., He C., Yan X., Mirshahi T., Logothetis D. E. (1999) Nat. Cell Biol. 1, 183–188 [DOI] [PubMed] [Google Scholar]
- 26.Guy-David L., Reuveny E. (2007) Neuron 55, 537–538 [DOI] [PubMed] [Google Scholar]
- 27.Du X., Zhang H., Lopes C., Mirshahi T., Rohacs T., Logothetis D. E. (2004) J. Biol. Chem. 279, 37271–37281 [DOI] [PubMed] [Google Scholar]
- 28.Milovic S., Steinecker-Frohnwieser B., Schreibmayer W., Weigl L. G. (2004) J. Biol. Chem. 279, 34240–34249 [DOI] [PubMed] [Google Scholar]
- 29.Liman E. R., Tytgat J., Hess P. (1992) Neuron 9, 861–871 [DOI] [PubMed] [Google Scholar]
- 30.Mirshahi T., Robillard L., Zhang H., Hébert T. E., Logothetis D. E. (2002) J. Biol. Chem. 277, 7348–7355 [DOI] [PubMed] [Google Scholar]
- 31.Banker G., Goslin K. (1998) Culturing Nerve Cells, 2nd Ed., pp. 339–370, The MIT Press, Cambridge, MA [Google Scholar]
- 32.Schreibmayer W., Lester H. A., Dascal N. (1994) Pflugers Arch. 426, 453–458 [DOI] [PubMed] [Google Scholar]
- 33.Lopes C. M., Franks N. P., Lieb W. R. (1998) Br. J. Pharmacol. 125, 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franks N. P., Lieb W. R. (1998) Toxicol. Lett. 100–101, 1–8 [DOI] [PubMed] [Google Scholar]
- 35.Eger E. I., 2nd, Fisher D. M., Dilger J. P., Sonner J. M., Evers A., Franks N. P., Harris R. A., Kendig J. J., Lieb W. R., Yamakura T. (2001) Anesthesiology 94, 915–921 [DOI] [PubMed] [Google Scholar]
- 36.Kulik A., Vida I., Fukazawa Y., Guetg N., Kasugai Y., Marker C. L., Rigato F., Bettler B., Wickman K., Frotscher M., Shigemoto R. (2006) J. Neurosci. 26, 4289–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jelacic T. M., Kennedy M. E., Wickman K., Clapham D. E. (2000) J. Biol. Chem. 275, 36211–36216 [DOI] [PubMed] [Google Scholar]
- 38.Koyrakh L., Luján R., Colón J., Karschin C., Kurachi Y., Karschin A., Wickman K. (2005) J. Neurosci. 25, 11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lüscher C., Slesinger P. A. (2010) Nat. Rev. Neurosci. 11, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuveny E., Slesinger P. A., Inglese J., Morales J. M., Iñiguez-Lluhi J. A., Lefkowitz R. J., Bourne H. R., Jan Y. N., Jan L. Y. (1994) Nature 370, 143–146 [DOI] [PubMed] [Google Scholar]
- 41.Carman C. V., Barak L. S., Chen C., Liu-Chen L. Y., Onorato J. J., Kennedy S. P., Caron M. G., Benovic J. L. (2000) J. Biol. Chem. 275, 10443–10452 [DOI] [PubMed] [Google Scholar]
- 42.He C., Zhang H., Mirshahi T., Logothetis D. E. (1999) J. Biol. Chem. 274, 12517–12524 [DOI] [PubMed] [Google Scholar]
- 43.Finley M., Arrabit C., Fowler C., Suen K. F., Slesinger P. A. (2004) J. Physiol. 555, 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He C., Yan X., Zhang H., Mirshahi T., Jin T., Huang A., Logothetis D. E. (2002) J. Biol. Chem. 277, 6088–6096 [DOI] [PubMed] [Google Scholar]
- 45.Ivanina T., Rishal I., Varon D., Mullner C., Frohnwieser-Steinecke B., Schreibmayer W., Dessauer C. W., Dascal N. (2003) J. Biol. Chem. 278, 29174–29183 [DOI] [PubMed] [Google Scholar]
- 46.Vivaudou M., Chan K. W., Sui J. L., Jan L. Y., Reuveny E., Logothetis D. E. (1997) J. Biol. Chem. 272, 31553–31560 [DOI] [PubMed] [Google Scholar]
- 47.Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishida M., MacKinnon R. (2002) Cell 111, 957–965 [DOI] [PubMed] [Google Scholar]
- 49.Nishida M., Cadene M., Chait B. T., MacKinnon R. (2007) EMBO J. 26, 4005–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin T., Peng L., Mirshahi T., Rohacs T., Chan K. W., Sanchez R., Logothetis D. E. (2002) Mol. Cell 10, 469–481 [DOI] [PubMed] [Google Scholar]
- 51.Yi B. A., Lin Y. F., Jan Y. N., Jan L. Y. (2001) Neuron 29, 657–667 [DOI] [PubMed] [Google Scholar]
- 52.Huang C. L., Jan Y. N., Jan L. Y. (1997) FEBS Lett. 405, 291–298 [DOI] [PubMed] [Google Scholar]
- 53.Sadja R., Alagem N., Reuveny E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10783–10788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X., Johnston D. (2005) J. Neurosci. 25, 3787–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sodickson D. L., Bean B. P. (1998) J. Neurosci. 18, 8153–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leaney J. L. (2003) Eur. J. Neurosci. 18, 2110–2118 [DOI] [PubMed] [Google Scholar]
- 57.Eger E. I., 2nd (2004) Am. J. Health Syst. Pharm. 61, Suppl. 4, S3–S10 [DOI] [PubMed] [Google Scholar]
- 58.Lazarenko R. M., Fortuna M. G., Shi Y., Mulkey D. K., Takakura A. C., Moreira T. S., Guyenet P. G., Bayliss D. A. (2010) J. Neurosci. 30, 9324–9334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marker C. L., Luján R., Colón J., Wickman K. (2006) J. Neurosci. 26, 12251–12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lerma J., Herranz A. S., Herreras O., Abraira V., Martín del Rio R. (1986) Brain Res. 384, 145–155 [DOI] [PubMed] [Google Scholar]
- 61.Bertaccini E. J., Trudell J. R., Franks N. P. (2007) Anesth. Analg. 104, 318–324 [DOI] [PubMed] [Google Scholar]
- 62.Sobolevsky A. I., Rosconi M. P., Gouaux E. (2009) Nature 462, 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X., Talley E. M., Patel N., Gomis A., McIntire W. E., Dong B., Viana F., Garrison J. C., Bayliss D. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3422–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.