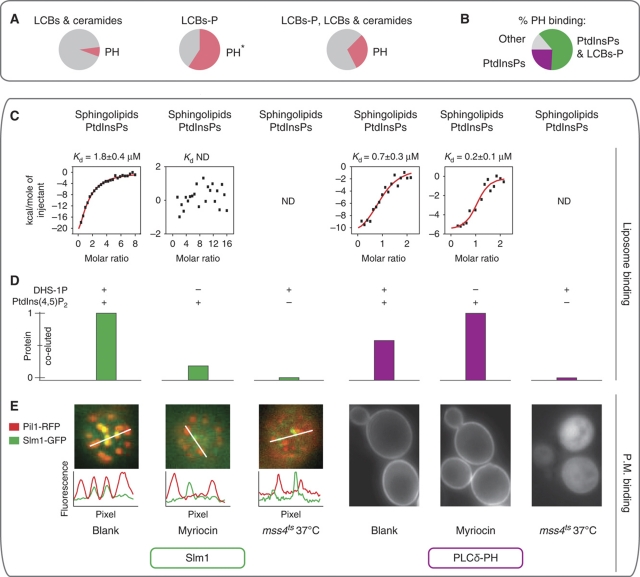
Figure 6.
The PH domain of Slm1 cooperatively binds PtdInsPs and phosphorylated sphingolipids. (A) A significant fraction of proteins-binding phosphorylated LCBs also contained a PH domain. For each sphingolipid-binding group, the pie chart shows the fraction of proteins with a PH domain (*P<0.00001). (B) Lipid-binding specificities of PH-domain containing proteins; more than half of those binding PtdInsPs also interacted with LCB-P. Proteins that did not show any binding were omitted. (C, D) Recruitment of Slm1-PH and PLCδ-PH to liposome membranes of different compositions: 3% (molar) PtdIns(4,5)P2 and/or 3% (molar) DHS-1P. (C) Quantitative measurement of liposome binding by isothermal titration calorimetry. Points represent heat evolved per mol of injected protein. ND: not determined. (D) Monitoring of Slm1-PH and PLCδ-PH co-eluting with liposomes by size exclusion chromatography and western blot. (E) Role of PtdInsPs and sphingolipids in the recruitment of Slm1 and PLCδ-PH to biological membranes in vivo. On the left, Slm1 fused to GFP (Slm1-GFP) co-localizes with eisosomes (Pil1-RFP). Interference with PtdIns(4,5)P2 metabolism using a temperature-sensitive mutant of the phosphatidylinositol 4-phosphate 5-kinase (mss4ts). Intensity plot profiles were extracted from the area represented by the respective white lines. On the right, localization of PLCδ-PH fused to GFP. Cell treatments are as in Figure 5A.