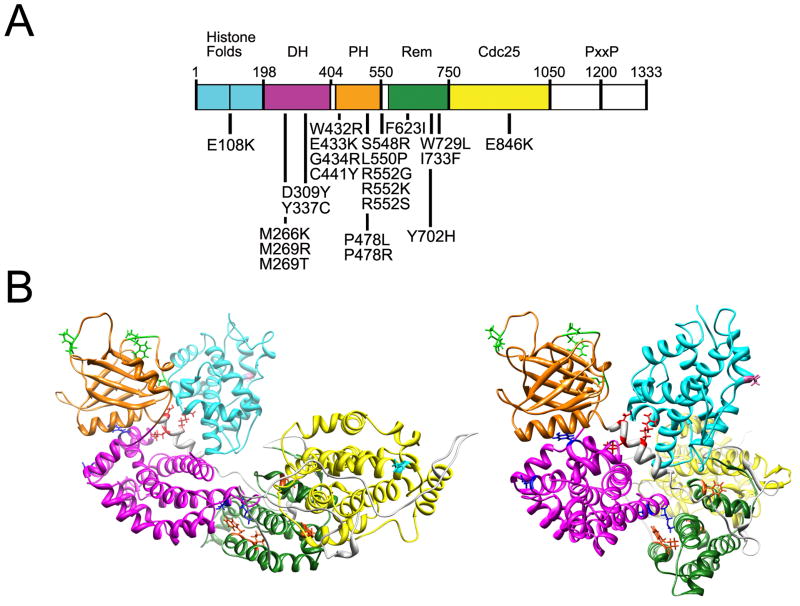
Figure 4. SOS1 domain structure and location of affected residues in NS.
(A) SOS1 missense mutations are positioned below the cartoon of the SOS1 protein with its functional domains indicated above. Abbreviations: DH, Dbl homology domain; PH, plekstrin homology domain; Rem, RAS exchanger motif; PxxP, proline-rich region. (B) Location of the mutated residues on the three-dimensional structure of SOS1. The functional domains are color coded as follows: Histone folds, cyan; DH, magenta; PH, orange; Rem, green; Cdc25, yellow. Residues affected by mutations are indicated with their lateral chains (histone folds, violet; DH, blue; PH, green; helical linker, red; Rem, orange; Cdc25, cyan). Based on Sondermann et al.121, which utilized structural data and computational modeling.