Fig. 1.
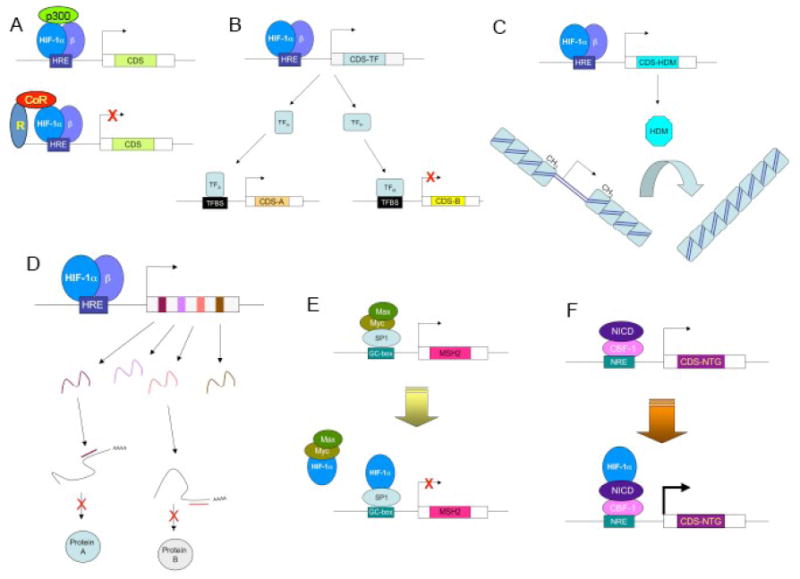
Mechanisms by which HIF-1 regulates gene expression. (A) Top: HIF-1 binds directly to target genes at a cis-acting hypoxia response element (HRE) and recruits coactivator proteins such as p300 to increase gene transcription. The coding sequence (CDS) of these HIF-1 target genes encode proteins that mediate adaptive responses to hypoxia. Bottom: There are only several rare examples in which HIF-1 binding leads to transcriptional repression. (B) HIF-1 binds directly to and transactivates a gene that encodes a transcription factor (TF), which either activates (TFA) or represses (TFB) the expression of secondary target genes by binding to its cognate transcription factor binding site (TFBS). (C) HIF-1 activates the transcription of genes encoding proteins with histone demethylase (HDM) activity. Demethylation of histones alters chromatin structure, which either decreases (as illustrated) or increases the expression of genes within the modified chromatin. (D) HIF-1 activates transcription of genes encoding microRNAs that bind to mRNAs and either block their translation or induce their degradation. (E) HIF-1a binds to the transcription factor SP1 and blocks activation of the gene encoding the mismatch DNA repair protein MSH2, thereby functioning as a co-repressor. (F) HIF-1a binds to the Notch intracellular domain (NICD) and potentiates transcriptional activation of Notch target genes, thereby functioning as a co-activator. (Adapted from ref. 50.)
