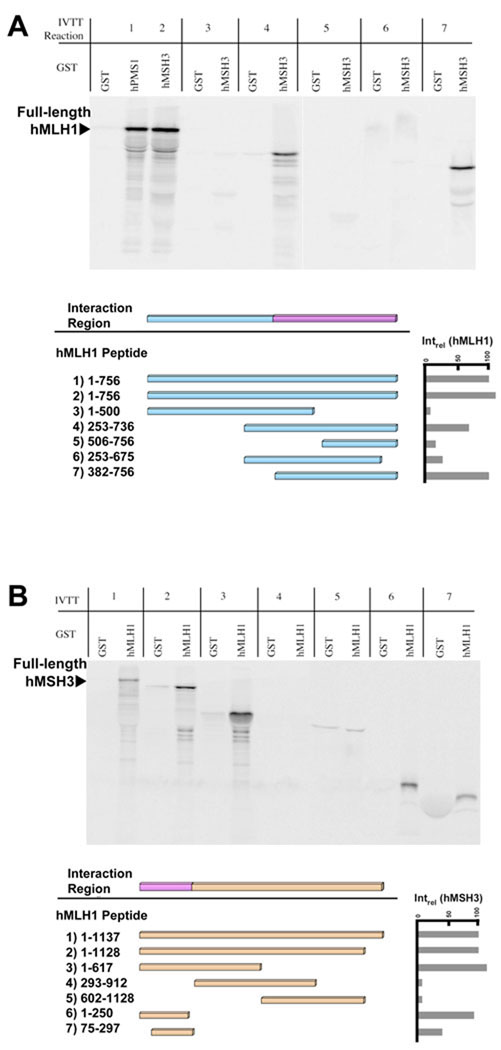
Fig. 5. Region of hMLH1 required for interaction with hMSH3.
(A) The region of hMLH1 required for its interaction with hMSH3 was determined via the GST interaction assay. Deletion constructs of hMLH1 were in vitro transcribed/translated and tested for interaction to GST-hMSH3. The interaction between hPMS1 and hMLH1 is a positive control and the reference for all other interactions(Intrel-hMLH1). Phosphorimager analysis was used for quantitation. The minimal region of hMLH1 required for interaction with hMSH3 consisted of amino acids 382–756 (lane 7). (B) The GST interaction assay was used to define the region of hMSH3 required for interaction with hMLH1. hMLH1 was tested against in vitro transcribed/translated deletion constructs of hMSH3. The interaction between hMLH1 and in vitro transcribed/translated hMLH3 served as a positive control and reference for all other interactions (Intrel-hMSH3). Phosphorimager analysis was used for quantitation. hMSH3 amino acids 1–250 is the minimal region required for interaction with hMLH1 (lane 6).