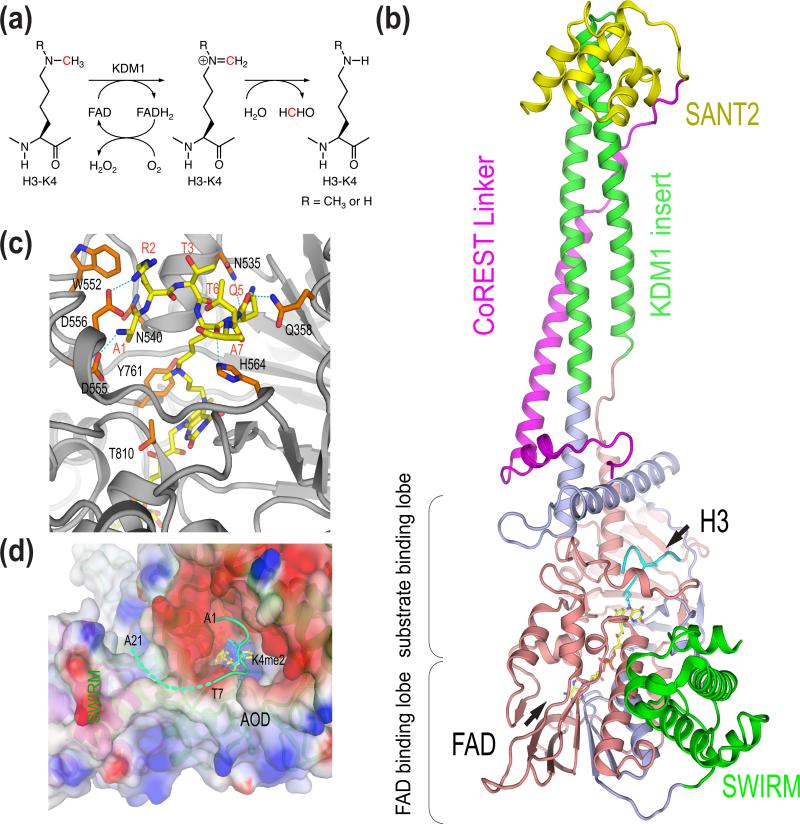
Figure 1.
Mechanism, structure, and substrate specificity of KDM1. (a) Chemical mechanism of KDM1-catalyzed lysine demethylation. (b) Structure of human KDM1-CoREST-H3 ternary complex (PDB code 2UXN). (c) Binding of H3 at the active site of KDM1. AOD is colored gray. The H3 peptide and residues in AOD involved in H3 binding are shown as sticks. (d) The potential involvement of the AOD-SWIRM interface in substrate binding. Residues 1-7 of H3 interact with the negatively charged active-site cavity of KDM1. The surface of AOD and SWIRM domain is colored by the electrostatic potential (red, negative; blue, positive). Residues 8-21 of H3 (shown as a dashed green line) and might bind at the groove between AOD and SWIRM.