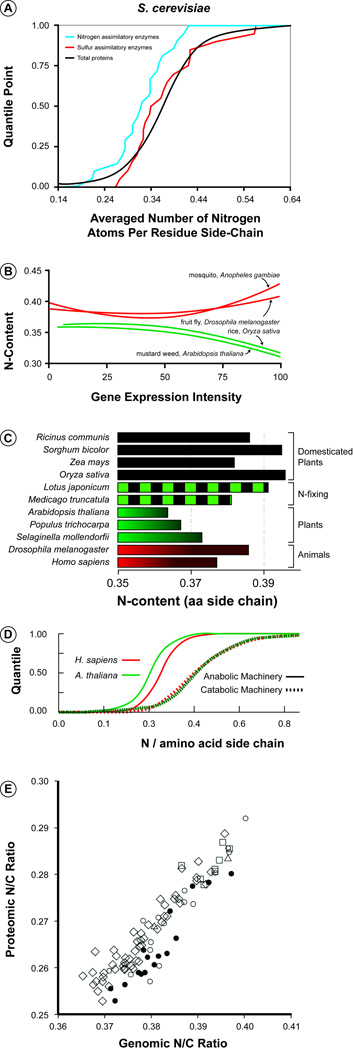
Figure 1.
Examples of element-based monomer usage bias (MUB) in prokaryotes and eukaryotes. (a) Nitrogen-processing enzymes (blue line) in yeast are unusually low in N compared to the average protein in the proteome (black) and to the average protein involved in sulfur-processing (red) [10], suggesting a form of “element-sparing” in molecules that are most important during limitation by a given element. (b) Protein N content declines with gene expression intensity in plants (green) but not in animals (red) [13], an outcome consistent with the fact that plants likely experience more frequent and sustained direct N limitation than do animals. (c) Proteome N-content in domesticated plants (black bars) and plants associated with N-fixing symbionts (green and black) is similar to that in animals (red) and higher in N-content than proteome N-content in undomesticated plants (green) [15], suggesting that release from selection pressures for N conservation has reduced MUB for N conservation in crop plants. Error bars (standard errors) are very small (max: ~0.001) and thus were not included, as they are not discernible on the plot. (d) The proteins involved in anabolic machinery (spliceosome and ribosomes) are higher in nitrogen content than are the proteins associated with catabolic machinery (proteosome, lysosome, and vacuole) for A. thaliana (green) and H. sapiens (red) [16], a result consistent with an interpretation of N-sparing during nutrient limitation when catabolic processes dominate. (e) In microorganisms, the N:C ratio of a taxon’s proteome is positively correlated with the N:C ratio of its genome [17], suggesting that selection for N conservation may even have been operating during early evolution of the canonical genetic code. Filled circles denote Archaea. Open symbols denote Eubacteria: free-living only, circle; capable of living as animal symbionts, diamond; capable of living as plant symbionts, square; capable of living as animal and plant symbionts, triangle.