Figure 1.
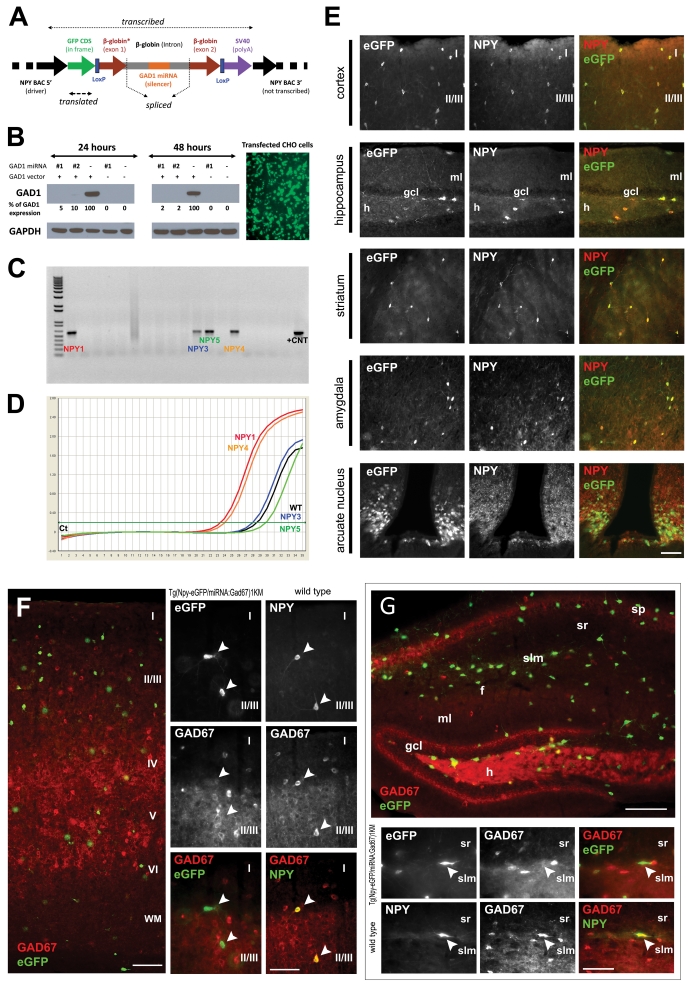
Generation and validation of the Tg(Npy-eGFP/miRNA:Gad1)1KM mice. (a) Schematic linear representation of the construct. Flanking regions of the construct are derived from the driver NPY BAC that provides interneuron subtype-specific expression of the construct. The 5′ region of the driver BAC (NPY promoter region) ensures specific expression of eGFP, and the SV40-pA ensures proper polyadenylation. The construct, containing a non-functional (not translated) part of β-globin exons 1 and 2, and intron 1 in its entirety, is spliced by the cellular machinery, liberating the intron that contains the 70–100 nucleotide long double-stranded miRNA directed against GAD1. The spliced intron-miRNA:Gad1 is processed through DROSHA and exported from the nucleus. In the cytoplasm the miRNA binds to and degrades the endogenous GAD1 mRNA through an RNA-induced silencing complex (RISC).33, 50 The eGFP mRNA is translated, thus fluorescently labeling cells of interest. The presence of LoxP sites facilitates generation of animals lacking the silencing part of the miRNA:Gad1 construct, but still expresses eGFP, to serve as controls. (b) miRNA-mediated downregulation of GAD1 in vitro. CHO cells co-transfected with GFP reporter vector alone (−), two different intermediate constructs (GFP reporter vectors containing two different synthetic miRNAs directed against GAD1—nos. 1 and 2), and GAD1 expression vector. The cells were harvested at 24 and 48 h after transfection. Western blot analysis confirmed that the two different miRNAs against GAD1 were correctly processed from the β-globin intron and that both miRNAs strongly downregulated GAD1 expression. Similar results have been obtained in HEK293 cells with stable expression of GAD1 that were engineered in our laboratory (data not shown). The same cells transfected with CMV-eGFP,miRNA:Gad1 show high levels of eGFP expression, confirming that the construct performs as expected. In addition, the processing of the miRNA from the β-globin intron in the construct does not interfere with eGFP protein translation. (c) Identification of founder animals by PCR-based genotyping of founder animals using eGFP primers with genomic DNA as template. The first lane represents size marker, and the last lane corresponds to positive control, with the remaining lanes containing eGFP amplification products from the genomic DNA of individual animals. The 550-nt product on a 1% agarose gel indicates construct incorporation into the genome of four founder animals (NPY 1, 3, 4 and 5). Similar results were obtained by Southern hybridization (data not sown). (d) Quantitative PCR (qPCR) amplification plot from frontal cortex of founder animals using eGFP construct-specific primers. The y axis denotes PCR product accumulation, and the x axis denotes amplification cycle number. Note that two (NPY1 and NPY4) of the four founder lines that incorporated the NPY-BAC/GAD1-miRNA construct reported functional eGFP expression. These lines were used for further characterization. (e) The NPY-BAC/GAD1-miRNA construct showed the expected tissue distribution in the brain. Micrographs depict the fidelity of co-localized eGFP and NPY in adult transgenic animals from line Tg(Npy-eGFP/miRNA:Gad1)1KM. The left column micrographs denote eGFP immunostaining, middle column micrographs represent sections labeled with anti-NPY and the right column micrographs illustrate pseudocolored composite of eGFP-NPY co-localization in the same tissue sections. In the cortex, roman numerals denote cortical laminae. Hippocampus abbreviations: gcl, granule cell layer; h, hilus; ml, molecular layer. Note that all GFP+ neurons are also NPY+, and all NPY+ cells are GFP+, suggesting that the construct is specifically and exclusively expressed in the phenotypically appropriate target cell population. Calibration bar=100 μm. (f, g) Tg(Npy-eGFP/ miRNA:Gad1)1KM animals show undetectable GAD1 levels in the frontal cortex and hippocampus of NPY+ cells compared with NPY+ neurons in control animals. eGFP-GAD1 double-immunohistochemistry (eGFP, green; GAD1, red) was performed from a coronal section through the frontal cortex and hippocampus of a transgenic animal. Wild-type control littermate was double stained against NPY and GAD1. Same cells are denoted by arrowheads. In the cortex, roman numerals denote cortical laminae. Hippocampal abbreviations: f, hippocampal fissure; gcl, ganglion cell layer; h, hilus; ml, molecular layer; slm, stratum lacunosum moleculare; sp, stratum pyramidale; sr, stratum radiatum. Note that all NPY+ cells (white arrows) are GAD1+ in control animals, whereas none of the eGFP+ (and thus NPY+) neurons (white arrows) show GAD1 staining in the transgenic mice. Moreover, note the large number of single-labeled GAD1+ neurons that are eGFP– in the transgenic line. These data indicate selective, miRNA-mediated, cell type-specific downregulation of GAD1 in NPY+ interneurons. Calibration bars=60 μm (overview figures) and 100 μm.