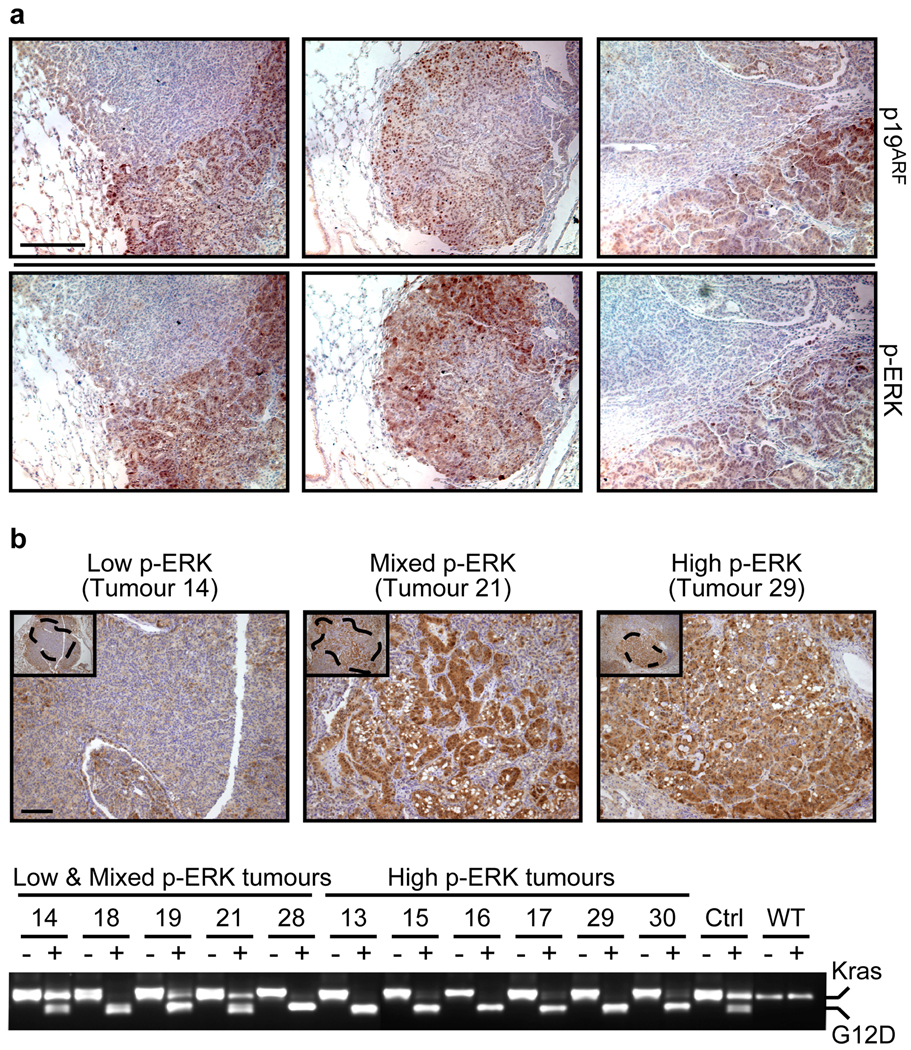
Figure 4. High-grade lung tumours exhibit increased Kras signalling.
a. IHC for p19ARF and p-ERK in consecutive sections of three independent low-to-high-grade transition tumours from KR;p53KI/KI mice. Scale bar = 200 µm.
b. Kras allele analysis was performed on genomic DNA from KR;p53KI/KI lung tumours following laser capture microdissection. p-ERK IHC was used to define areas of low, mixed or high p-ERK (upper panel, Scale bar=50µm) and consecutive slides used for LCM of defined regions (see dotted areas). DNA was isolated from LCM material and the Kras genomic region amplified by PCR and digested with HindIII (lower panel). For each tumour, the undigested (−) and digested (+) PCR fragments were run alongside and the wt (Kras, higher band) and mutant alleles (G12D, lower band) are indicated. Control lung tissue from heterozygous (KrasG12D/+: Ctrl) and wild-type (WT) mice was also analyzed.