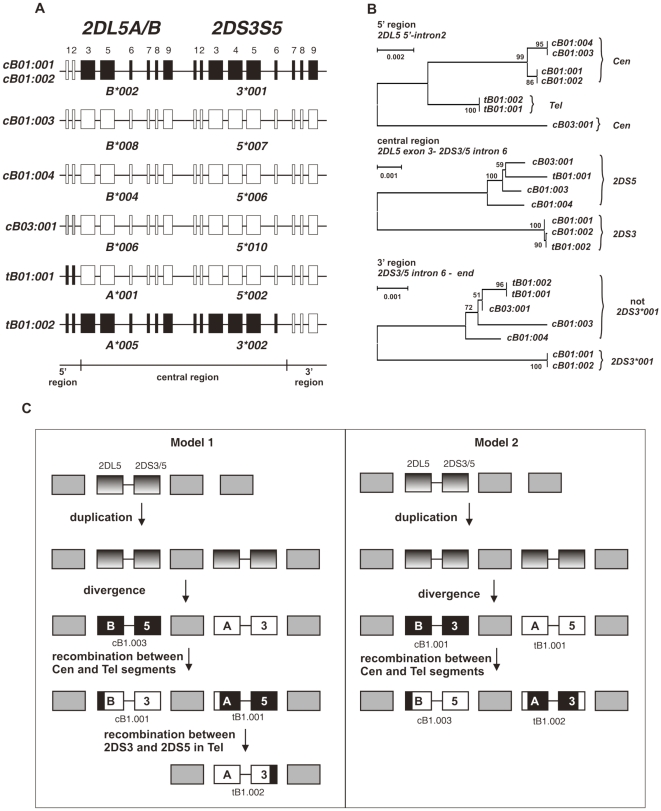
Figure 5. 2DS3 and 2DS5 descend from a common ancestor and diversified by interlocus recombination.
(A) Shows the six combinations of 2DL5 and 2DS3/5 present in the 27 haplotypes, along with the motif to which they belong. Shading denotes their relatedness as determined by the phylogenetic analyses shown in panel B. (B) Genomic sequences extending from 5′ of 2DL5 through the end of the neighboring 2DS3/5 gene were aligned, divided into three regions (5′, central, and 3′) and submitted to phylogenetic analysis. Neighbor joining trees are shown, with Bootstrap values for the nodes. For the 5′ region (from 250 bp 5′ of 2DL5 exon 1 through to the end of intron 2), the variants divide according to their location in the centromeric or telomeric half of the KIR locus. The central region (from exon 3 of 2DL5 to 1.9 kb 5′ of exon 6 in 2DS3S5) divides according to association with 2DS3 or 2DS5. In the 3′ segment (from intron 6 to the polyadenylation signal at the end of 2DS3S5) the variants containing 2DS3*001 form an outgroup. (C) Shows two models for the evolution of 2DS3 and 2DS5. Both models start with a single 2DL5A/B-2DS3/5 progenitor in the centromeric region, the only site where chimpanzee and orangutan 2DL5 has been found ([65] and AC220148). This progenitor then duplicated, with one daughter being transposed to the telomeric region. Subsequent diversification at the two loci resulted in distinct 2DL5A, 2DL5B, 2DS3 and 2DS5 genes. The two models differ in the sites where 2DS3 and 2DS5 arose: in model 1, 2DS5 arose in the centromeric site, 2DS3 in the telomeric site, whereas in model 2, 2DS3 arose in the centromeric site, 2DS5 in the telomeric site. Subsequent recombination between variants in the centromeric and telomeric sites gave rise to the variants observed in the modern KIR haplotypes. Model 2 requires only two such recombinations and is more parsimonious than model 1, which requires three. However, the greater diversity observed in 2DL5B-2DS5 than 2DL5A-2DS5, favors model 1 over model 2.