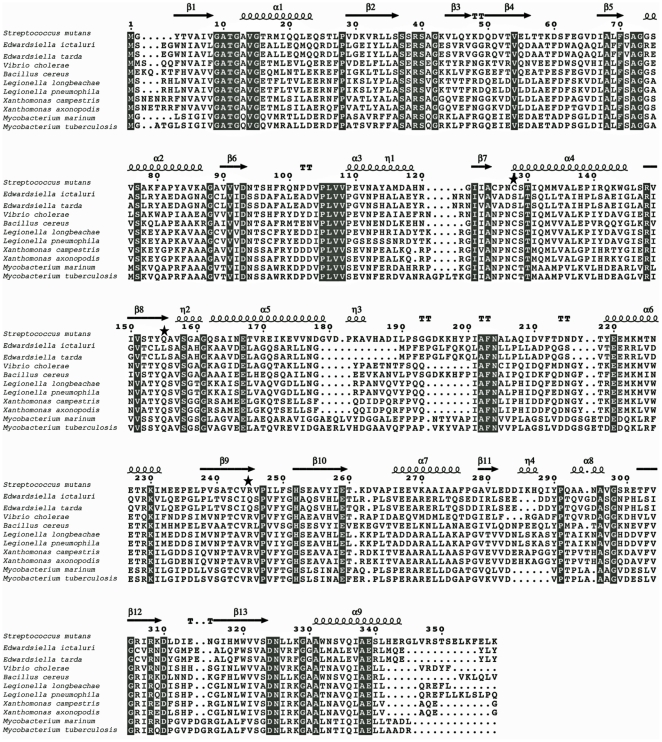
Figure 2. Sequence alignment among representative members of the AsdB family.
The secondary structure at the top of the alignment corresponds to the S. mutans AsdB enzyme (spirals represent α-helix; arrows represent β-sheet). Conserved amino acids residues are indicated in grey. The stars indicated the key catalytic active site residues not present in AsdB from Edwardsiella. The AsdB sequences were obtained from NCBI's Entrez Protein database for Streptococcus mutans NP_721384.1; Edwardsiella ictaluri YP_002934124; Edwardsiella tarda YP_003296462; Vibrio cholerae YP_001217630.1; Bacillus cereus YP_085142.1; Legionella longbeachae CBJ10915; Legionella pneumophila YP_096311.1; Xanthomonas axonopodis NP_643032.1; Xanthomonas campestris NP_637897.1; Mycobacterium tuberculosis NP_218225.1; Mycobacterium marinum YP_001853481.1.