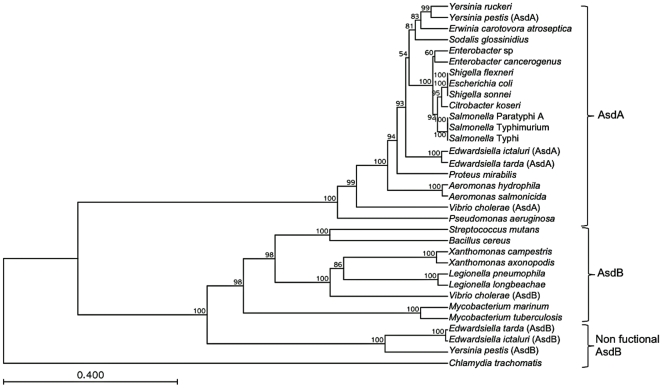
Figure 3. Phylogenetic tree constructed by the unweighted pair group method with arithmetic mean.
Bootstrap values indicate the number of times that a given node was detected out of 100. The Asd sequences were obtained from NCBI's Entrez Protein database for Edwardsiella ictaluri YP_002935083.1; Edwardsiella tarda YP_003297386.1; Escherichia coli AP_004358.1; Salmonella Typhi NP_807591.1; Salmonella Paratyphi A YP_152515.1; Salmonella Typhimurium AAB69392.1; Shigella flexnieri YP_690789.1; Shigella sonnei YP_312455.1; Citrobacter koseri YP_001456333.1; Enterobacter cancerogenus ZP_05969786.1; Enterobacter sp. YP_001178547.1; Yersinia pestis NP_671174.1; Yersinia ruckeri ZP_04615435.1; Proteus mirabilis YP_002152826.1; Aeromonas hydrophila ABK39477.1; Aeromonas salmonicida YP_001142146.1; Sodalis glossinidius YP_456010.1; Vibrio cholerae YP_002810714.1; Pseudomonas aeruginosa NP_251807.1; Erwinia carovora atrosepticum YP_052242.1; Streptococcus mutans NP_721384.1; Edwardsiella ictaluri YP_002934124; Edwardsiella tarda YP_003296462; Vibrio cholerae YP_001217630.1; Bacillus cereus YP_085142.1; Legionella longbeachae CBJ10915; Legionella pneumophila YP_096311.1; Xanthomonas axonopodis NP_643032.1; Xanthomonas campestris NP_637897.1; Mycobacterium tuberculosis NP_218225.1; Mycobacterium marinum YP_001853481.1; Chlamydia trachomatis YP_002887982.1.