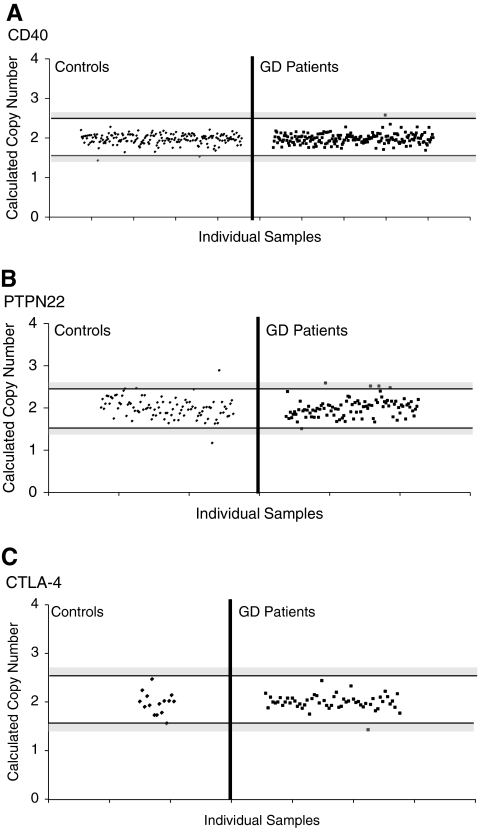
FIG. 2.
Copy number variation analysis of CD40, PTPN22, and CTLA-4. TaqMan CNV assays were selected for CD40 and PTPN22 covering CNVs currently deposited in the Database of Genomic Variation. For CTLA-4, however, there was no cataloged CNV in the database, so a CNV assay encompassing the 5′UTR of the CTLA-4 gene was chosen. Each dot represents the copy number of each individual sample. The area between the horizontal black bars represents the range of values that constitutes two copies. Gray-shaded regions signify borderline values that cannot be clearly assigned, either a one or two copies (lower region) or a two or three copies (upper region). Samples that fell in these gray areas were excluded from analysis. Calculated copy numbers of CD40 (A), PTPN22 (B), and CTLA-4 (C) in control and Graves' disease (GD) patient samples are shown. CD40 and CTLA-4 showed no copy number variations in either the controls or the GD patients. PTPN22 showed no variation in the GD population, and only rare variation, one duplication and one deletion, in the control group.