Abstract
The voltage-dependent anion channel (VDAC), located in the outer mitochondrial membrane, acts as a gatekeeper for the entry and exit of mitochondrial metabolites. Here we reveal functional dynamics of isoform one of VDAC (VDAC1) by a combination of solution NMR spectroscopy, Gaussian network model analysis, and molecular dynamics simulation. Micro- to millisecond dynamics are significantly increased for the N-terminal six β-strands of VDAC1 in micellar solution, in agreement with increased B-factors observed in the same region in the bicellar crystal structure of VDAC1. Molecular dynamics simulations reveal that a charge on the membrane-facing glutamic acid 73 (E73) accounts for the elevation of N-terminal protein dynamics as well as a thinning of the nearby membrane. Mutation or chemical modification of E73 strongly reduces the micro- to millisecond dynamics in solution. Because E73 is necessary for hexokinase-I-induced VDAC channel closure and inhibition of apoptosis, our results imply that micro- to millisecond dynamics in the N-terminal part of the barrel are essential for VDAC interaction and gating.
Keywords: membrane protein, molecular dynamics, NMR spectroscopy, protein-lipid interactions, structure
Dynamics of proteins provide the essential link between structure and function. Membrane protein dynamics are gaining more attention due to an increasing number of high-resolution structures. A versatile experimental method for the study of membrane protein dynamics is NMR spectroscopy, providing atomic resolution information from nanoseconds to seconds (for an overview see ref. 1). In addition, when a three-dimensional structure is available, insight into fast dynamics of proteins, which occur on the nanosecond time scale, might be gained from molecular dynamics (MD) simulation and elastic network models (2, 3).
NMR studies have revealed large conformational changes essential for the physiological function of α-helical membrane proteins, such as the interaction of phospholamban with effectors modulating heart muscle contractility (4, 5) and antimicrobial peptides adopting different conformations in membranes or solution (6, 7). Less well characterized are motions of outer membrane β-barrels. A pioneering solution NMR study revealed μs-ms interconversion of the catalytic loop of PagP between an excited and a nonexcited state, enabling both ligand binding and enzymatic catalysis (8). Nonenzymatic β-barrel channels exhibit more general dynamic features, such as loop flexibility and slow conformational exchange towards the extracellular barrel edges, altogether indicating relevance for substrate conductance and gating (9–11).
The voltage-dependent anion channel (VDAC) is one of the few β-barrel membrane proteins, the structure of which has been determined to date (12–14). Serving as the major pass-through for metabolites between mitochondria and the cytosol (15), the highly abundant channel is regarded as a key regulator of cellular energy metabolism (16). Metabolite flow is regulated by a voltage-dependent conformational change between a high-conductance anion-selective state, several low-conductance states favoring small cations (17, 18), and a high-conductance cation-selective state (19). VDAC gating is furthermore modulated by a variety of ions, small molecules, apoptotic proteins of the Bcl-2 family, and hexokinase (20), strongly implicating VDAC’s function in apoptosis regulation (21). To enable interactions and gating between various states, conformational variability of VDAC is expected. Indeed, large structural rearrangements have been proposed on the basis of electron microscopy (22) and bilayer measurements (23). However, high-resolution structural information about these conformational changes is missing.
Here we characterized the dynamics of VDAC1 by a combination of solution NMR, Gaussian network model (GNM) analysis, and MD simulation. We show that extensive μs-ms dynamics occur in the N-terminal β-strands of VDAC1, and that these motions are reduced by removal of the charge on E73 by either mutation or chemical modification. In addition, our MD simulations demonstrate that the charge on the side chain of E73 facing the membrane in the barrel is not only causing enhanced dynamics of the barrel wall, but at the same time results in thinning of the nearby membrane. Our findings link the dynamics of VDAC to the channel’s function and show the involvement of a large barrel region and a membrane-facing charged residue in functional dynamics of a β-barrel membrane protein.
Results
μs-ms Dynamics Are Significantly Increased for the N-Terminal Six β-Strands of VDAC1.
Previously, we had assigned 70% of all backbone resonances of human VDAC1 (hVDAC1) corresponding to 90% of residues within the barrel (12). With improved sample quality and additional NMR experiments we now increased the assignment to 84/97% (overall/barrel). The assignment of three residues in the N-terminal helix (K12, D16, and V17) benefited from the chemical shift information obtained from solid-state NMR (24). In total, five residues are assigned in the second half of the N-terminal helix (residues 12–20), demonstrating that this region is not broadened beyond detection by intermediate chemical exchange. This result is in line with the observation that peaks corresponding to residues A2–V17 give rise to distinct narrow crosspeaks in solid state NMR spectra of hVDAC1 reconstituted in liposomes, indicating a rather rigid conformation (24).
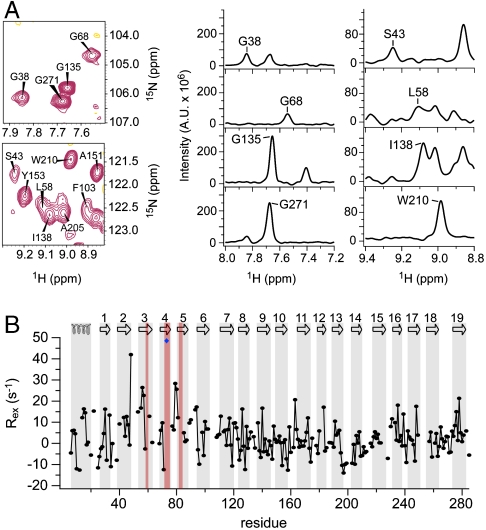
Next we characterized the backbone dynamics of hVDAC1 using NMR spectroscopy. Amide protons in the N-terminal four β-strands of wild type (WT) hVDAC1 exhibit fast hydrogen/deuterium (H/D) exchange, pointing to conformational exchange that occurs on a time scale of minutes or faster (12). In addition, average peak intensities observed in two-dimensional 1H,15N-TROSY spectra for residues located in the N-terminal six β-strands were about 1.5-fold lower than those of the central β-strands (β7-β15) (Fig. 1A and Fig. S1A). The C-terminal strands β16–β19 were also broadened (Fig. S1A), but broadening in this region intensified with sample age and therefore has to be considered with caution. In contrast, lower signal intensities in the N-terminal six β-strands were observed consistently for many samples with different age. The low signal intensities complicated the resonance assignment of the N-terminal β-strands and the signals of some N-terminal barrel residues could not be identified. Lower NMR signal intensities can be a consequence of either increased rigidity in the ps-ns range or increased conformational exchange that is intermediate on the NMR time scale (μs-ms dynamics).
Fig. 1.
μs-ms dynamics are significantly increased for the N-terminal β-strands of VDAC1. (A) Enlarged spectral regions and one-dimensional traces of a 1H,15N-TROSY spectrum of hVDAC1 measured at a 1H frequency of 900 MHz showing examples of broadened and nonbroadened peaks. (B) Chemical exchange rates (Rex) measured at 900 MHz. The topology of VDAC1 is indicated at the top, with secondary structure elements highlighted in gray. Residues that could not be assigned are highlighted in red; E73 is indicated by a blue diamond.
To probe for the influence of oligomerization on the observed signal broadening we measured a two-dimensional 1H,15N-TROSY spectrum at a fivefold lower concentration. Despite the fivefold lower concentration the strong signal broadening in the N-terminal β-strands remained (Fig. S2). In addition, the N-terminal four β-strands of WT hVDAC1 exhibit fast H/D exchange (12), whereas oligomerization would be expected to reduce H/D exchange rates in the interacting region. Thus, oligomerization is not a major factor responsible for the observed signal broadening in the N-terminal β-strands of hVDAC1. Further contributions to signal broadening may come from remote influences caused by motions of nearby groups (side chains or detergent molecules), rather than by motions of the barrel residues themselves. Again, these influences do not explain the fast H/D exchange (12).
To determine the time scale of dynamics causing the NMR signal broadening in the N terminus of hVDAC1, we measured heteronuclear NOE (HetNOE) values (25) and 15N transverse spin relaxation rates (26). HetNOE values, which have a field-dependent maximum value of around 0.7–0.9 for a fully rigid backbone, are sensitive to dynamics on the ps-ns time scale while 15N transverse spin relaxation rates probe chemical exchange occurring in the μs-ms range. HetNOE values in different hVDAC1 β-strands were similar with an average value of 0.75 (Fig. S1B). Slightly smaller HetNOE values were observed for strands β2 and β3. In addition, the HetNOE values decreased towards the edges of the strands and in the loops (Fig. S1B), pointing to increased flexibility on the ps-ns time scale. Residues with increased flexibility on the ps-ns time scale are expected to result in more intense NMR signals, in contrast to what was observed in hVDAC1 (Fig. 1A and Fig. S1A). Thus, the combination of NMR signal intensities with HetNOE measurements pointed to the presence of chemical exchange that is intermediate on the NMR time scale.
Next we directly measured chemical exchange contributions to 15N spin relaxation rates (26). Although chemical exchange rates (Rex) were distributed around ∼0 s-1 with a rather large standard deviation, three regions of elevated exchange were determined (Fig. 1B and Fig. S1C): Residues located in β2–β7, β16–17, and β19 showed average values ≥3.5 s-1. β19 exhibited increased values especially in the first half of the strand, which might be attributed to a mismatch in hydrogen bonding to the shorter strand β1. In addition, the parallel alignment of β1 and β19 might result in a lower intrinsic stability and thus favor the conformational exchange observed in β19. The most pronounced chemical exchange was present in the N-terminal barrel ranging from β2 to β7 with the largest average Rex values seen in β3 for K53 (14.9 s-1), T55 (16.7 s-1), G56 (26.7 s-1), S57 (22.7 s-1), and K61 (12.8 s-1), corresponding to time scales of 37 to 78 ms. Nine residues in β3–β5 could not be assigned due to overlap or chemical exchange broadening. In addition, several residues in the N-terminal barrel (e.g., K34, T42, S43, G45, and S57) showed two distinct resonances in two-dimensional 1H,15N-TROSY spectra and, in the case of S57, in three-dimensional sequential correlation TROSY-1H,15N, 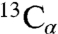 (TROSY-HNCA) spectra (Fig. S3), directly proving the existence of a second conformation. Thus, strong μs-ms conformational exchange is present in the N-terminal part of the hVDAC1 barrel centered at strands β3–β4.
(TROSY-HNCA) spectra (Fig. S3), directly proving the existence of a second conformation. Thus, strong μs-ms conformational exchange is present in the N-terminal part of the hVDAC1 barrel centered at strands β3–β4.
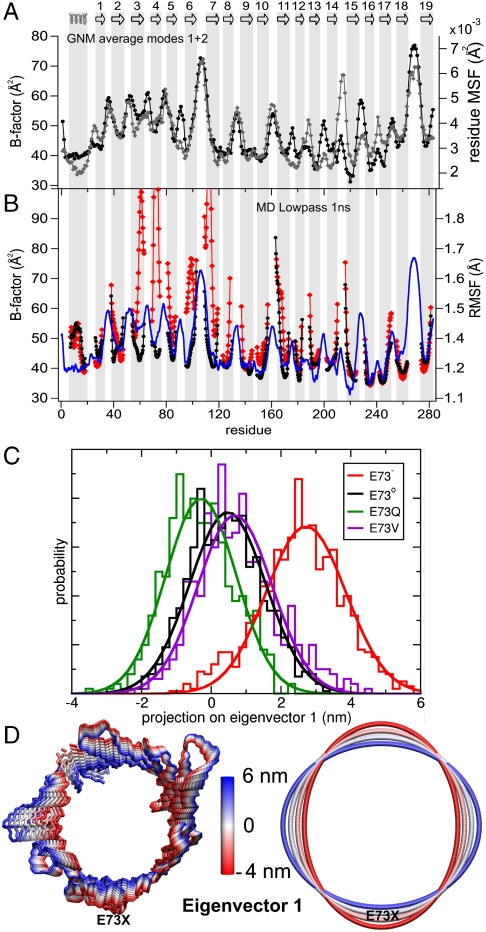
Increased B-Factors in the Crystal Structure of VDAC Correlate with Low-Frequency Modes Derived from Gaussian Network Model Analysis.
Dynamic features of proteins are also present in crystals, partly captured by Debye-Waller factors or B-factors. B-factors measure both thermal motions of atoms (dynamic disorder) and small differences in the structure of individual molecules (static disorder) (27). VDAC1 from mouse (mVDAC1) was crystallized in a bicellar environment (14). mVDAC1 differs from human VDAC1 by just four amino acid substitutions, namely T55N, M129V, A160S, and I227V. The three-dimensional structures of human and murine VDAC1 are almost identical (12, 14). In the crystal structure of mVDAC1 the B-factors for residues in the β-strands are highly nonuniform (Fig. 2A). Whereas in the central strands average B-factors of 41.3 Å2 were observed, β2–β7 and β18–β19 showed average values of 50.6 Å2 and 48.9 Å2, respectively.
Fig. 2.
A charge on the membrane-facing E73 accounts for the elevation of N-terminal protein dynamics of VDAC1. (A) Comparison of crystallographic B-factors (black circles) of mVDAC1 (PDB code 3EMN) with B-factors predicted by the average mobility of the two lowest frequency modes obtained from the GNM analysis (gray diamonds). (B) rmsF of the backbone of mVDAC1 embedded in a DMPC phospholipid bilayer observed during a 150 ns MD simulation. The curves correspond to low-pass filtered (1 ns) rmsF values of the VDAC1 E73- (red) and E73o (black) simulations, respectively. The loop residues were not included. The blue curve shows the crystallographic B-factors of mVDAC1 (3EMN). The topology of mVDAC1 is sketched at the top, with numbered β-strands displayed as arrows. (C) PCA projection along the first eigenvector of mVDAC1, generated from the combined MD trajectories of all four E73 variants, using the backbone atoms of the β-barrel from residue 26 to 283. Normalized histograms and Gaussian fits of the PCA projection show the extent of space of eigenvector 1 spanned by E73- (red), E73o (black), E73Q (green), and E73V (purple) trajectories. (D) Interpolation of the structure of mVDAC1 along the first PCA eigenmode using a backbone representation (left) and an elliptic fitting (right) of the Cα atoms of the backbone representation. The two extremes correspond to the two extremes in the histograms of E73- (+6 nm) and E73o (-4 nm) in (C).
Crystallographic B-factors can be predicted from a three-dimensional structure by a diverse range of methods. We used GNM (28), an isotropic, Cα-atom connected elastic network model, to test if the unusually high B-factors of the N-terminal β-strands are related to the intrinsic stability of the three-dimensional structure. While B-factors predicted from the 20 lowest frequency modes in the GNM analysis yielded low correlations with crystallographic B-factors, the usage of the average of the two lowest frequency modes resulted in a notable agreement between crystallographic and GNM-predicted B-factors (Fig. 2A), with an overall correlation coefficient of R2 = 0.61 and a value of R2 = 0.58 for β-strands only, which is substantial regarding the complexity of the prediction. Motions with low frequency are believed to be associated with high cooperativity, suggesting that the high crystallographic B-factors and GNM-predicted low-frequency fluctuations show a predisposition of the N-terminal barrel region for its involvement in collective motions. Mapping of B-factors and GNM-derived residue fluctuations onto the crystal structure showed that β-strands next to the helix (the central strands β8–β15) were highly ordered, ascribing a stabilizing function to the N-terminal helix (residues 6–20) (Fig. S4). Such a stabilizing role of the helix has also been suggested on the basis of solid-state NMR results on an N-terminally truncated version of hVDAC1 (24). In contrast, high flexibility was found from β2 to the top end of β7 (Fig. S4). Thus, the same regions that are strongly affected by chemical exchange on the μs-ms time scale have an increased disorder in the crystal structure, in agreement with a lower intrinsic stability predicted from the two lowest eigenmodes of the GNM analysis.
VDAC Flexibility from MD Simulations and the Role of Charge on E73.
Elastic network models like the GNM analysis derive protein dynamics directly from the three-dimensional structure by modeling the protein atoms (in this case only Cα-atoms) within a defined distance range as Hookean springs (2, 3). The exact time scale of dynamics, the effect of point mutations (except for changes in the size of a side chain), and the influence of the membrane environment can thus not be investigated by the GNM analysis. To overcome these limitations, we performed MD simulations of mVDAC1 inserted into a dimyristoylphosphatidylcholine (DMPC) phospholipid bilayer covering simulation times up to ∼150 ns. MD simulations were performed for both WT mVDAC1 and mutated forms, which probe the importance of the charge and polarity of the side chain of E73: the deprotonated charged (E73-) and the protonated uncharged (E73o) WT mVDAC1, and further the polar (E73Q) and hydrophobic (E73V) mutant forms. From the root-mean-square fluctuations (rmsF) observed in the MD simulations B-factors were calculated. In addition, to extract motions slower or faster than 1 ns, a low-pass and high-pass filter, respectively, were employed. Our analysis showed that in the fast regime (< 1 ns) the mobility of mVDAC1 was only marginally influenced by the charge state of E73 (Fig. S5). In contrast, fluctuations occurring on time scales above 1 ns significantly increased from the uncharged to the charged form of E73 (Fig. 2B). The rise in mobility was especially pronounced between β-strands 2–7.
To track global changes in the β-barrel, a principal component analysis (PCA) of the backbone dynamics was performed. A one-dimensional projection of the WT and mutant mVDAC1 trajectories onto the PCA eigenvector 1 (EV1) (Fig. 2C) demonstrates that E73o and both neutral E73 mutants sampled a similar region of PCA space along the first eigenmode. In contrast, a charge on the side chain of E73 resulted in a larger coverage of space toward higher projections along EV1 (Fig. 2C). Eigenvector 1 describes the main differences between WT and mutants, including differences in the average structure. A representation of the motion along the first eigenmode on the β-barrel and a fit of this mode to an ellipse (Fig. 2D) showed that this mode can be viewed primarily as a global elliptic deformation of the barrel structure around E73.
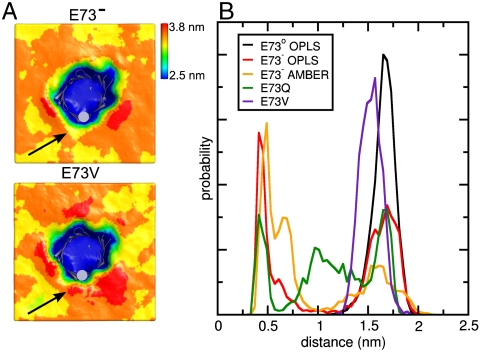
A Negative Charge on the Side Chain of E73 Leads to Thinning of the Nearby Membrane.
To further assess changes caused by mutations or protonation of E73, we analyzed perturbations of the lipid bilayer in the MD simulations. Comparison of the average bilayer thickness in the case of E73- and E73V (Fig. 3A) demonstrates that a negative charge on the side chain carboxylate group of E73 leads to a marked thinning of the membrane around this residue. When the carboxylate group of E73 is uncharged by protonation (E73o) no thinning occurs, while the polar, but uncharged E73Q mutant displayed thinning to a lower extent (Fig. S6A), indicating that the commonly used replacement of glutamic acid with glutamine cannot fully mimic the uncharged form of E73 in our case. We therefore attribute membrane deformation primarily to the presence of a charge at position 73.
Fig. 3.
Membrane perturbation due to charge on E73. (A) Average thickness of DMPC bilayers calculated from MD simulations with E73- (top) or E73V (bottom) mVDAC1 inserted. The secondary structure of mVDAC1 is shown as gray cartoons and mVDAC1 position 73 as gray sphere. Arrows indicate the influenced membrane areas next to E73 and E73V. (B) Normalized minimum distance distribution between choline nitrogen atoms and mVDAC1 position 73. Probabilities are given in arbitrary units.
The distortion of the membrane frequently correlated with a flipping motion of lipids in the bilayer, causing a transient approach of the phospholipid head groups towards E73 (Fig. S6B). The flipping motion was reflected in an altered distribution of the distance between E73 and the closest lipid head group (Fig. 3B). As can be seen, E73- and E73Q substantially promoted the approach of lipid head groups from a distance of ∼1.5 nm to ∼0.5 nm to group 73, i.e., into the central region of the membrane.
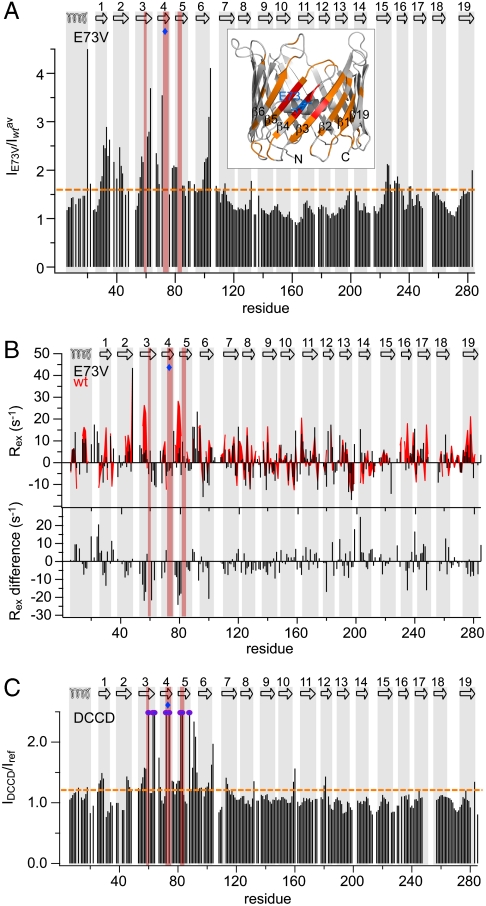
The Single-Point Mutation E73V and the Reaction of E73 with DCCD Stabilize the VDAC Barrel.
Functional studies have ascribed an important function to E73, showing that ruthenium red and hexokinase-I mediated gating and apoptosis inhibition are impaired in E73Q VDAC (29). Moreover, we previously showed that E73V hVDAC1 exhibits reduced H/D-exchange in the N-terminal barrel region (12), indicating that motion on a second to minute time scale responsible for H/D exchange is strongly quenched. To dissect the influence of E73 on hVDAC1 dynamics in solution, we analyzed NMR signal intensities for the single-point-mutation E73V. Strikingly, NMR signal intensities were strongly increased in the N terminus in E73V hVDAC1 compared to the wild-type protein (Fig. 4A). The changes were centered around E73 involving a large area of the barrel (Fig. 4A inset). In the case of WT hVDAC1 the peak broadening was shown to be caused by chemical exchange (Fig. 1B and Fig. S1C), indicating that the mutation E73V strongly reduces the μs-ms time scale dynamics in the N-terminal barrel region. Indeed, a comparison of relaxation rates of E73V hVDAC1 with WT values (Fig. 4B) demonstrates a reduction of μs-ms time scale dynamics for β1–β9 and β19, with the largest changes in β3 and β5 as well as the loop connecting β4 and β5.
Fig. 4.
Stabilization of hVDAC1 by the E73V mutation and by DCCD reaction. (A) Ratio of peak intensities between E73V and WT hVDAC1 observed in 1H,15N-spectra, averaged over a three-residue window. The orange dashed line indicates the threshold value of 1.6. Residues with a larger ratio were mapped onto the three-dimensional structure (PDB code 2JK4) in orange (inset). Residues that, in contrast to E73V hVDAC1, could not be assigned in WT hVDAC1 are mapped in red. (B) Top: chemical exchange rates (Rex) of E73V (black bars) compared to WT values from Fig. 1B (red area). Difference values (E73V— WT) are shown below. (C) DCCD-induced changes in resonance intensities observed in 1H,15N-TROSY spectra of hVDAC1, averaged over a three-residue window. Purple circles indicate residues that show up after DCCD treatment. The orange dashed line is the threshold value of 1.2. (A–C) The topology of hVDAC1 is indicated at the top, showing the location of the N-terminal helix and the β-strands. Residues that, in contrast to E73V hVDAC1, could not be assigned in WT hVDAC1 are highlighted in red, and a blue diamond marks the position of E73.
E73 can be modified by reaction with N,N′-dicyclohexylcarbodiimide (DCCD), a chemical that reacts with aspartic and glutamic acid residues in hydrophobic surroundings. DCCD has previously been shown to specifically bind VDAC1 in mitochondrial membranes at position E73 (30). The specificity of the reaction is also given in micellar solution, because chemical shift changes upon DCCD addition were localized close to E73 (Fig. S7A). Importantly, reaction with DCCD resulted in increased peak intensities for many residues (Fig. 4C). In addition, most of the amide resonances that are missing in WT hVDAC1 spectra but are present in E73V hVDAC1 spectra, appeared, e.g., T60, R63, W64, T72, K74, G82, T83, and E88 (Fig. 4C and Fig. S7 B and C). No changes in NMR signal position or intensity were observed in the C-terminal region upon reaction with DCCD in contrast to the finding for E73V hVDAC1 (compare Fig. 4A).
Discussion
Increasing evidence suggests that conformational changes in VDAC are important for (voltage-dependent) gating and induction of apoptosis (as reviewed in ref. 31). Large conformational changes during gating have long been proposed by electron microscopy (22) and bilayer measurements (23). Recent studies propose that these motions involve a reorientation of the N-terminal helix (14, 32). The modulation of VDAC channel conductance by small molecules and apoptotic proteins in vivo (20) strongly implicates VDAC as a regulator of apoptosis (21). Various models for the channel’s regulatory part exist, some proposing that VDAC is part of the permeability transition pore responsible for the release of apoptogenic factors from the mitochondria into the cytosol upon apoptotic stimuli (31).
By a combination of solution NMR spectroscopy, Gaussian network model analysis, and MD simulation we showed that (i) μs-ms dynamics are significantly increased for the N-terminal six β-strands of VDAC1, (ii) a charge on the membrane-facing E73 plays a key role for the elevation of N-terminal protein dynamics, (iii) mutation and chemical modification of E73 strongly reduces μs-ms dynamics in solution, and (iv) charge on the side chain of E73 facing the membrane in the barrel is coupled to thinning of the nearby membrane. The dynamics of VDAC1 presented in this study set the protein apart from bacterial β-barrel membrane proteins. While for bacterial porins loops play a major role in gating or catalysis (8–11, 33, 34), our study reveals a unique involvement of a large area of the β-barrel in functional dynamics of a eukaryotic membrane protein.
NMR spectroscopy revealed conformational exchange, which occurs in the N-terminal β-strands β2–β7 of the hVDAC1 barrel on the time scale of μs-ms (Fig. 1). In addition to slow dynamics, increased ps-ns flexibility in lauryldimethylamine-N-oxide (LDAO)-solubilized hVDAC1 was detected for strands β2 and β3 by NMR spectroscopy (Fig. S1B). The MD simulations of VDAC1 embedded in lipid bilayers were performed for simulation times up to ∼150 ns and can thus probe protein dynamics that occur on the ns time scale. B-factors in crystal structures have been found to correlate well with NMR derived S2 order parameters reporting on ps-ns dynamics (35, 36). In addition, static disorder contributions to B-factors may reflect slower dynamics when multiple conformations are present in the crystal. In extreme cases of conformational exchange, electron densities are often not refined [e.g., in CitA (37)], such that possible correlations of B-factors and slow dynamics are invisible. In the case of the N-terminal region of mVDAC1, contributions to B-factors might arise from dynamics and static disorder intermediate in amplitude such that crystal structure refinement was possible. Together the data indicate the presence of extensive protein dynamics in the N-terminal part of the VDAC1 barrel on a wide range of time scales. In addition, as the flexibility of the N-terminal part of the VDAC1 barrel is increased on time scales above 1 ns in the MD simulations (Fig. 2B), the correlation with crystallographic B-factors was best for the lowest frequency GNM modes (Fig. 2A), and strong chemical exchange in the μs-ms time scale was observed by NMR spectroscopy (Fig. 1B), slow collective motions are likely to play a key role. Part of these slow collective motions could be the global deformation of the VDAC1 barrel that was observed in the MD simulations (Fig. 2D). Experimental information about these slow motions might also be gained from Carr-Purcill-Meiboom-Gill (CPMG) NMR relaxation dispersion experiments (38), which can be used to characterize the structure of low-populated excited states, which are invisible with other techniques. Although a TROSY-CPMG version for large molecules exists (39), the very high molecular weight in the case of the VDAC-micelle complex likely exceeds the limit for recording high quality data required to draw reliable conclusions about the structure of the excited state(s). An indication for the structure of the excited state comes from the second conformation of S57 in the three-dimensional TROSY-HNCA spectrum. The Cα chemical shift value of the second conformation of S57 was shifted by ∼0.2 ppm towards the random coil value (Fig. S3B), suggesting that the minor conformation is less structured.
Our study demonstrates that the dynamics in the barrel of hVDAC1 are mainly determined by the membrane-facing residue E73. While in micellar solution as well as the crystal structure from bicelles the protonation state of E73 is unclear, the MD simulations clearly show that a charge on the side chain of E73 has the ability to increase motion in the N-terminal barrel and to reduce the membrane thickness next to E73. Other functionally relevant glutamic and aspartic acids, which are located in hydrophobic pockets of proteins, have pKa values as high as 8 (40). Assuming a high pKa value for VDAC as well, VDAC is partially protonated in solution at the pH used here (6.8). Furthermore, the reaction with DCCD requires the presence of carboxylate (41), indirectly proving the existence of a (partial) charge in solution. DCCD inhibits the channel activity of VDAC1 in a voltage-dependent manner, requiring incubation at high negative or positive potentials. In addition, DCCD slows down the transition from the high-conducting to a long-lived low-conducting state by stabilizing intermediate states of the VDAC1 channel (42). Occupation of long-lived intermediate states is in agreement with the reduction in intrinsic dynamics by DCCD modification reported here. We conclude that the decreased protein dynamics observed in the N terminus of hVDAC1 upon mutation of E73 to valine and by chemical modification with DCCD is caused by removal of the partial charge.
Our observations have important implications for VDAC1’s function as a gatekeeper for metabolites and its role in apoptosis. Residues suggested to be involved in gating of Saccharomyces cerevisiae VDAC are located in the N-terminal helix, the linker, as well as the first five β-strands (43). E73, located within this region, is conserved in human VDAC isoform 2 (hVDAC2) and in mammals. Mutation of E73 to glutamine (E73Q) shows reduced voltage-dependence of gating and inhibits ruthenium red (RuR)- and hexokinase-mediated effects on channel function and protection from apoptosis (29). The observed effects were attributed to inefficient binding of hexokinase to the mutated form of VDAC (44). Moreover, the reaction of VDAC1 with DCCD abolishes the interaction of VDAC1 with hexokinase (29, 45). Thus, our results imply that dynamics in the N-terminal region are relevant for hexokinase binding. This interpretation is in line with recent findings showing destabilized regions of outer membrane proteins to be preferentially involved in protein-protein interaction (46).
Our MD simulations of mVDAC1 showed that the nature and charge status of a single residue, in this case E73, can be highly coupled to the global motion of a protein. This result is remarkable, as E73 is not located anywhere near a hinge region but roughly in the middle of a barrel-forming β-strand. At the same time the charge and polarity of E73 is strongly correlated with the dynamics and conformation of the membrane environment, suggesting that the dynamics of the lipids might be connected with global protein motions. This finding is interesting as lipid-protein interactions are known to be important for the structure and function of integral membrane proteins (47–52). In particular, VDAC1 gating and ionic selectivity have been shown to be dependent on lipid composition (53). Our MD simulations of mVDAC1 in a lipid bilayer demonstrated that a charge on E73 causes a perturbation of the surrounding membrane (Fig. 3). Because the major lipid components of outer mitochondrial membranes are positive lipids (phosphatidylcholine and phosphatidylethanolamine) (54), the membrane perturbation is likely to occur also in vivo and might relate to nonspecific membrane rupture proposed to occur during apoptosis (55).
Materials and Methods
Expression, refolding, and purification of WT and E73V hVDAC1 was done as described previously (12). For chemical modification of hVDAC1, a fourfold molar excess of DCCD (Calbiochem-Novabiochem) was added. NMR spectra were recorded on 2H(75%)-15N or 2H(75%)-13C-15N labeled samples containing 0.5–0.8 mM hVDAC1, 25 mM BisTris pH 6.8, approximately 250 mM LDAO (Fluka), and 5–10% D2O. Improvement of backbone assignment was achieved by TROSY-HNCA experiments (56), 1H,15N-TROSY experiments (57) at varying temperatures and Cα shifts from solid-state NMR experiments (24). Backbone dynamics were determined from steady state heteronuclear {1H},15N-NOEs (25) and chemical exchange rates (Rex) (26).
Normal modes from the mVDAC1 crystal structure were predicted by GNM analysis (28). MD simulation in DMPC lipids was performed for 100–150 ns using the crystal structure of mVDAC1 (14) and modified versions thereof as starting structures. Further details on the methods are provided as SI Text.
Supplementary Material
Acknowledgments.
We thank M. Bayrhuber and K. Zeth for help in the initial stage of the project and V. Gapsys for porting GridMAT-MD algorithms to GROMACS. This work was supported by the Max Planck Society, the Fonds der Chemischen Industrie (scholarship to S.V.), the Deutsche Forschungsgesellschaft (DFG) [Emmy Noether Fellowship to A.L. and Sonderforschungsbereich (SFB) 803], and through a DFG Heisenberg scholarship (ZW 71/2-2 to M.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012310108/-/DCSupplemental.
References
- 1.Veglia G, Ramamoorthy A. Special issue on “membrane protein dynamics: correlating structure to function”. Biochim Biophys Acta. 2010;1798:65–67. doi: 10.1016/j.bbamem.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys Rev Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 3.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 4.Traaseth NJ, Veglia G. Probing excited states and activation energy for the integral membrane protein phospholamban by NMR CPMG relaxation dispersion experiments. Biochim Biophys Acta. 2010;1798:77–81. doi: 10.1016/j.bbamem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu S, Coey AT, Lorigan GA. Solid-state (2)H and (15)N NMR studies of side-chain and backbone dynamics of phospholamban in lipid bilayers: investigation of the N27A mutation. Biochim Biophys Acta. 2010;1798:210–215. doi: 10.1016/j.bbamem.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjya S, Ramamoorthy A. Multifunctional host defense peptides: functional and mechanistic insights from NMR structures of potent antimicrobial peptides. FEBS J. 2009;276:6465–6473. doi: 10.1111/j.1742-4658.2009.07357.x. [DOI] [PubMed] [Google Scholar]
- 7.Salnikov E, Aisenbrey C, Vidovic V, Bechinger B. Solid-state NMR approaches to measure topological equilibria and dynamics of membrane polypeptides. Biochim Biophys Acta. 2010;1798:258–265. doi: 10.1016/j.bbamem.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Hwang PM, Kay LE. Solution structure and dynamics of integral membrane proteins by NMR: a case study involving the enzyme PagP. Methods Enzymol. 2005;394:335–350. doi: 10.1016/S0076-6879(05)94013-5. [DOI] [PubMed] [Google Scholar]
- 9.Liang B, Arora A, Tamm LK. Fast-time scale dynamics of outer membrane protein A by extended model-free analysis of NMR relaxation data. Biochim Biophys Acta. 2010;1798:68–76. doi: 10.1016/j.bbamem.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 11.Renault M, et al. Solution state NMR structure and dynamics of KpOmpA, a 210 residue transmembrane domain possessing a high potential for immunological applications. J Mol Biol. 2009;385:117–130. doi: 10.1016/j.jmb.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 12.Bayrhuber M, et al. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiller S, et al. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ujwal R, et al. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benz R. Permeation of hydrophilic solutes through mitochondrial outer membranes: review on mitochondrial porins. Biochim Biophys Acta. 1994;1197:167–196. doi: 10.1016/0304-4157(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 16.Lemasters JJ, Holmuhamedov E. Voltage-dependent anion channel (VDAC) as mitochondrial governator—thinking outside the box. Biochim Biophys Acta. 2006;1762:181–190. doi: 10.1016/j.bbadis.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Schein SJ, Colombini M, Finkelstein A. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from paramecium mitochondria. J Membr Biol. 1976;30:99–120. doi: 10.1007/BF01869662. [DOI] [PubMed] [Google Scholar]
- 18.Blachly-Dyson E, Forte M. VDAC channels. IUBMB Life. 2001;52:113–118. doi: 10.1080/15216540152845902. [DOI] [PubMed] [Google Scholar]
- 19.Pavlov E, et al. The mitochondrial channel VDAC has a cation-selective open state. Biochim Biophys Acta. 2005;1710:96–102. doi: 10.1016/j.bbabio.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS. The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Curr Pharm Design. 2006;12:2249–2270. doi: 10.2174/138161206777585111. [DOI] [PubMed] [Google Scholar]
- 21.Shoshan-Barmatz V, Keinan N, Zaid H. Uncovering the role of VDAC in the regulation of cell life and death. J Bioenerg Biomembr. 2008;40:183–191. doi: 10.1007/s10863-008-9147-9. [DOI] [PubMed] [Google Scholar]
- 22.Guo XW, Mannella CA. Conformational change in the mitochondrial channel, VDAC, detected by electron cryo-microscopy. Biophys J. 1993;64:545–549. doi: 10.1016/S0006-3495(93)81399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmerberg J, Parsegian VA. Polymer inaccessible volume changes during opening and closing of a voltage-dependent ionic channel. Nature. 1986;323:36–39. doi: 10.1038/323036a0. [DOI] [PubMed] [Google Scholar]
- 24.Schneider R, et al. The native conformation of the human VDAC1 N terminus. Angew Chem Int Ed Engl. 2010;49:1882–1885. doi: 10.1002/anie.200906241. [DOI] [PubMed] [Google Scholar]
- 25.Zhu G, Xia Y, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Rance M, Palmer AG., III Mapping chemical exchange in proteins with MW > 50 kD. J Am Chem Soc. 2003;125:8968–8969. doi: 10.1021/ja035139z. [DOI] [PubMed] [Google Scholar]
- 27.Ringe D, Petsko GA. Study of protein dynamics by X-ray diffraction. Methods Enzymol. 1986;131:389–433. doi: 10.1016/0076-6879(86)31050-4. [DOI] [PubMed] [Google Scholar]
- 28.Yang LW, et al. oGNM: online computation of structural dynamics using the Gaussian Network Model. Nucleic Acids Res. 2006;34:W24–31. doi: 10.1093/nar/gkl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaid H, Abu-Hamad S, Israelson A, Nathan I, Shoshan-Barmatz V. The voltage-dependent anion channel-1 modulates apoptotic cell death. Cell Death Differ. 2005;12:751–760. doi: 10.1038/sj.cdd.4401599. [DOI] [PubMed] [Google Scholar]
- 30.De Pinto V, al Jamal JA, Palmieri F. Location of the dicyclohexylcarbodiimide-reactive glutamate residue in the bovine heart mitochondrial porin. J Biol Chem. 1993;268:12977–12982. [PubMed] [Google Scholar]
- 31.Shoshan-Barmatz V, et al. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med. 2010;31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Hiller S, Wagner G. The role of solution NMR in the structure determinations of VDAC-1 and other membrane proteins. Curr Opin Struct Biol. 2009;19:396–401. doi: 10.1016/j.sbi.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yildiz O, Vinothkumar KR, Goswami P, Kuhlbrandt W. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 2006;25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong H, Szabo G, Tamm LK. Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat Chem Biol. 2006;2:627–635. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- 35.Clore GM, Schwieters CD. Concordance of residual dipolar couplings, backbone order parameters and crystallographic B-factors for a small alpha/beta protein: a unified picture of high probability, fast atomic motions in proteins. J Mol Biol. 2006;355:879–886. doi: 10.1016/j.jmb.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Tasayco ML, Polenova T. Dynamics of reassembled thioredoxin studied by magic angle spinning NMR: snapshots from different time scales. J Am Chem Soc. 2009;131:13690–13702. doi: 10.1021/ja9037802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sevvana M, et al. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J Mol Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 38.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill Sequence for characterizing chemical exchange by NMR Spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 39.Loria JP, Rance M, Palmer AG., 3rd A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J Biomol NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 40.Valiyaveetil F, Hermolin J, Fillingame RH. pH dependent inactivation of solubilized F1F0 ATP synthase by dicyclohexylcarbodiimide: pK(a) of detergent unmasked aspartyl-61 in Escherichia coli subunit c. Biochim Biophys Acta. 2002;1553:296–301. doi: 10.1016/s0005-2728(01)00251-1. [DOI] [PubMed] [Google Scholar]
- 41.Khorana HG. The chemistry of carbodiimides. British Columbia Research Council. 1953:145–166. [Google Scholar]
- 42.Shafir I, Feng W, Shoshan-Barmatz V. Dicyclohexylcarbodiimide interaction with the voltage-dependent anion channel from sarcoplasmic reticulum. Eur J Biochem. 1998;253:627–636. doi: 10.1046/j.1432-1327.1998.2530627.x. [DOI] [PubMed] [Google Scholar]
- 43.Thomas L, Blachly-Dyson E, Colombini M, Forte M. Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc Natl Acad Sci USA. 1993;90:5446–5449. doi: 10.1073/pnas.90.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abu-Hamad S, Zaid H, Israelson A, Nahon E, Shoshan-Barmatz V. Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: mapping the site of binding. J Biol Chem. 2008;283:13482–13490. doi: 10.1074/jbc.M708216200. [DOI] [PubMed] [Google Scholar]
- 45.Nakashima RA, Mangan PS, Colombini M, Pedersen PL. Hexokinase receptor complex in hepatoma mitochondria: evidence from N,N′-dicyclohexylcarbodiimide-labeling studies for the involvement of the pore-forming protein VDAC. Biochemistry. 1986;25:1015–1021. doi: 10.1021/bi00353a010. [DOI] [PubMed] [Google Scholar]
- 46.Naveed H, Jackups R, II, Liang J. Predicting weakly stable regions, oligomerization state, and protein-protein interfaces in transmembrane domains of outer membrane proteins. Proc Natl Acad Sci USA. 2009;106:12735–12740. doi: 10.1073/pnas.0902169106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nyholm TK, Ozdirekcan S, Killian JA. How protein transmembrane segments sense the lipid environment. Biochemistry. 2007;46:1457–1465. doi: 10.1021/bi061941c. [DOI] [PubMed] [Google Scholar]
- 48.Langosch D, Arkin IT. Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 2009;18:1343–1358. doi: 10.1002/pro.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kagan BL, Thundimadathil J. Amyloid peptide pores and the beta sheet conformation. Adv Exp Med Biol. 2010;677:150–167. doi: 10.1007/978-1-4419-6327-7_13. [DOI] [PubMed] [Google Scholar]
- 50.Galdiero S, Galdiero M, Pedone C. beta-Barrel membrane bacterial proteins: structure, function, assembly and interaction with lipids. Curr Protein Pept Sci. 2007;8:63–82. doi: 10.2174/138920307779941541. [DOI] [PubMed] [Google Scholar]
- 51.Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc Natl Acad Sci USA. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunte C, Richers S. Lipids and membrane protein structures. Curr Opin Struct Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Rostovtseva TK, Bezrukov SM. VDAC regulation: role of cytosolic proteins and mitochondrial lipids. J Bioenerg Biomembr. 2008;40:163–170. doi: 10.1007/s10863-008-9145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta. 1997;1325:108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- 55.Rostovtseva TK, Tan W, Colombini M. On the role of VDAC in apoptosis: fact and fiction. J Bioenerg Biomembr. 2005;37:129–142. doi: 10.1007/s10863-005-6566-8. [DOI] [PubMed] [Google Scholar]
- 56.Eletsky A, Kienhofer A, Pervushin K. TROSY NMR with partially deuterated proteins. J Biomol NMR. 2001;20:177–180. doi: 10.1023/a:1011265430149. [DOI] [PubMed] [Google Scholar]
- 57.Zhu G, Kong XM, Sze KH. Gradient and sensitivity enhancement of 2D TROSY with water flip-back, 3D NOESY-TROSY and TOCSY-TROSY experiments. J Biomol NMR. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.