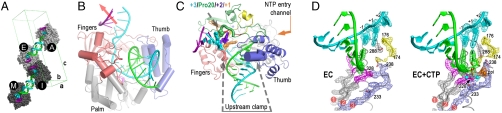
Fig. 1.
Poliovirus 3Dpol elongation complex. (A) Crystal packing showing staggered coaxial stacking of upstream template-product duplexes resulting in two nonequivalent pairs of ECs (A&E vs. I&M). Product strand is shown in green, template in cyan, and downstream nontemplate in purple. (B) EC structure showing up- and downstream RNA duplexes as the template strand (cyan) threads through the active site and the red arrow indicates trajectory of downstream RNA duplex. (C) Top view of EC showing the single stranded conformation of the +1, +2, and +3 downstream template nucleotides and the protein clamp of the upstream duplex. Palm domain is in gray, thumb is in blue, and the individual fingers (19) are colored with index in green, middle in orange, ring in yellow, and the pinky in pink. (D) 3,500 K composite simulated-annealing omit maps contoured at 1.5 σ showing quality of active site electron density for the native (EC) and Mg2+-CTP (EC + CTP) complexes. The pyrophosphate (ppi) in the CTP complex is shown in orange and the presence of metals ions was confirmed by an essentially identical Mn2+-CTP structure (see Figs. S3 and S4).