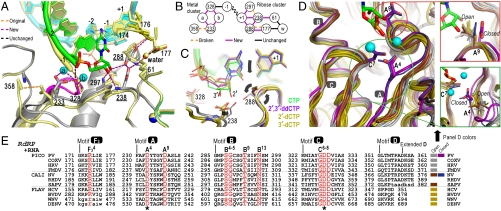
Fig. 3.
Molecular interactions involved in RdRP active site closure. (A) Superposition of 3Dpol active site in open state 1 (yellow) and closed state 4 (colors) showing how ribose hydroxyl recognition drives active site closure and divalent metal binding via the two distinct clusters of interactions diagrammed in panel (B). (C) Comparison of bound CTP, 2′-dCTP, 3′-dCTP, and 2′,3′-ddCTP conformations illustrating the NTP movement associated with active site closure. Only CTP with both ribose hydroxyls triggers the movements (arrows) of Asp238A9 and Ser288B4 that result in the closed conformation and catalysis. β and γ phosphates are omitted for clarity. (D) Conservation of motif A open conformation among all positive-strand RNA virus polymerases shown by superposition of four picornaviral (gray), two caliciviral (olive), and four flaviviral (brown) polymerases and a direct comparison with the closed conformation polio (purple) and Norwalk (blue) virus polymerase structures. Mg2+ ions and newly incorporated CMP are from the state 4 structure and the expanded views show the conformations of key side chains in all the structures. Note that a rabbit hemorrhagic disease virus polymerase structure adopting an intermediate conformation induced by Lu3+ binding is not shown. (E) Structure-based sequence alignment of RdRP motifs A–D and F3. Residues playing key roles in active site interactions and closure are colored red, the two invariant catalytic Asp residues are highlighted by asterisks, and residues in lower case letters either deviate from the consensus structure conformations or are not resolved in the crystal structures and are therefore included based only on sequence homology. See Fig. S8A for an extended alignment of all polymerase motifs across all classes of single-subunit polymerases and Fig. S8B for PDB codes.