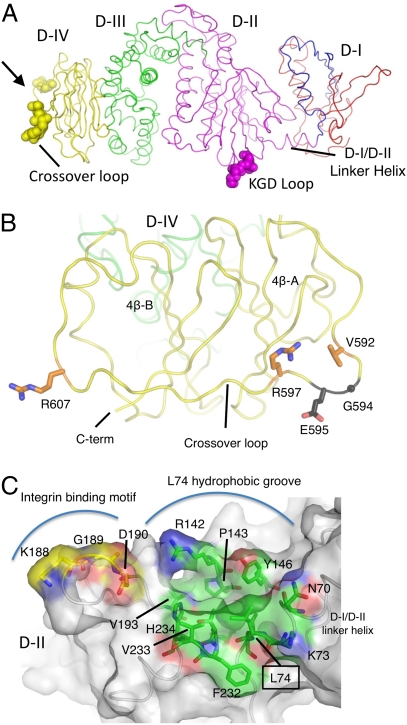
Fig. 4.
Residues in gH D-II to D-IV implicated in EBV entry. (A) Residues in EBV gH D-II and D-IV that are implicated in EBV entry are shown on the structure as CPK spheres, colored by domain as in Fig. 1. The D-IV residues are located at the extreme C-terminal end of the molecule in a cross-over loop between the two halves of the domain. The D-II residues, defining an integrin binding motif implicated in epithelial cell entry, are located in D-II, near the D-I/D-II linker helix. (B) gH residues G594 and E595 are located in the D-IV cross-over loop connecting the two β-sheets of the domain fold. The view shown is oriented ∼90° about the vertical axis from the view in A (bold arrow). The residues are shown as dark gray sticks with O and N atoms colored red and blue, respectively. gH residues 592, 597, and 607, which have moderate effects on gH/gL fusion activity, are also exposed on the D-IV cross-over loop. These residues are shown as orange sticks, with O and N atoms colored red and blue. (C) The gH/gL integrin binding site is adjacent to an extended hydrophobic groove including L74. The view shown is oriented ∼90° about the horizontal axis from the view in A. The gH/gL chain is shown as a coil beneath a transparent protein surface. The KGD motif is shown as sticks with carbon atoms colored yellow and mapped onto the gH/gL surface. The nearby residue L74, located in the D-I/D-II linker helix, is surrounded by gH residues that form a primarily hydrophobic groove lined with charged residues. These are also shown in stick representation, labeled, and colored with carbon atoms in green.