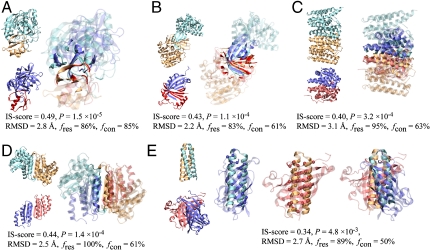
Fig. 2.
Examples of similar protein–protein interface pairs identified by iAlign. Coordinates of structures were taken from the PDB. The template (cyan/orange) and target (blue/red) proteins are (A) subtilisin BPN/chymotypsin inhibitor 2 (PDB code and chain IDs: 1tm7_EI) and streptogrisin B/ovomucoid inhibitor (1sgy_EI), (B) ribokinase (1vm7_AB) and heme-degrading enzyme PC130 (1sqe_AB), (C) farnesyl pyrophosphate synthetase (1rtr_AB) and HemAT (1or6_AB), (D) aspartate racemase (1jfl_AB) and DCoH (1dcp_EF), and (E) Rop (1f4m_CD) and allene oxide cyclase (1z8k_AC). In E, interfacial alignments are illustrated separately for each side of the interface; and the Cα atoms of aligned residues are represented in spheres. For clarity, interface/noninterface regions are shown in solid/transparent colors, respectively. Molecular images were created with VMD (32).