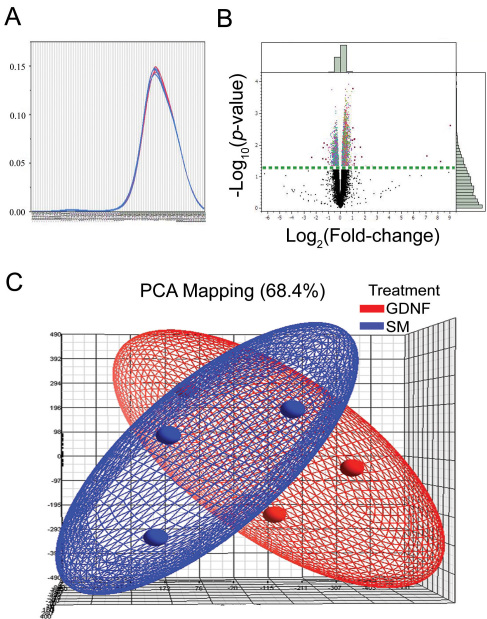
Figure 7.
Overlayed kernel density estimates, principle component analysis, and volcano plot. A: Overlayed kernel density estimates show the raw univariate distributions for all arrays. The distributions for these arrays are very similar, indicating that this is a high-quality data set that should require little if any normalization for further analysis. B: Volcano plot from ANOVA analysis of the differential expression under epidermal growth factor (EGF) and EGF+glial cell line-derived neurotrophic factor (GDNF) conditions reveals differentially expressed genes. The fold change values between two conditions (log10 transformed) were plotted on the x-axis and were compared to the negative log10 transformed p values on the y-axis. Genes with a transformed p value of at least 0.05 and a transformed fold change of at least two in the upper left and upper right of the volcano plot are highlighted in red. Among these genes, eight are predicted genes and the other 16 are listed in Table 2. Histograms along the borders were generated via all detected genes. The dotted straight horizontal line represents the value 1.3 [-log10 (p value=0.05)], and the genes above that line are significantly different in expression level between the two conditions. The pale green bars in the histograms along the borders show numbers of all genes, and the dark green bars inside represent genes with a p value of <0.05. C: Principle component analysis (PCA) of gene expression signals reveals a broad degree of overlap in expression patterns between the two treatment conditions, with little suggestion of a treatment effect. Blue indicates data from retinal progenitor cells (RPCs) cultured in standard, epidermal growth factor-based medium (SM), while red indicates data from standard medium supplemented with GDNF. Individual microarrays (3 each condition) are indicated as small orbs and the general data distribution for each condition as an elliptical field of the corresponding color.