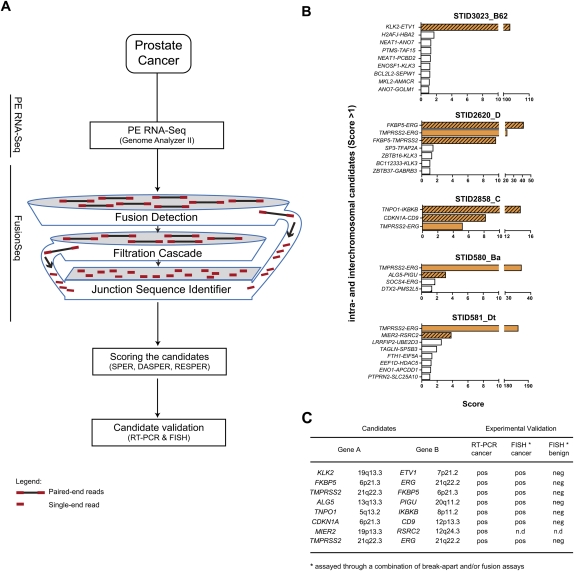
Figure 1.
FusionSeq identifies seven high-scoring intra- and interchromosomal candidates that could be validated positively in five prostate cancer samples. (A) Schematic of the computational processes composing FusionSeq. (B) List of candidates within five samples. Candidates with negative DASPER scores were removed, and the remaining candidates were sorted anticlimactic by RESPER (referred to as Score) with a cutoff of 1. True gene fusions (solid orange, known TMPRSS2–ERG fusions; striated orange, novel gene fusions) score higher than candidates that appear down the list (white bars). (C) The novel gene fusions were experimentally validated using RT-PCR and FISH.