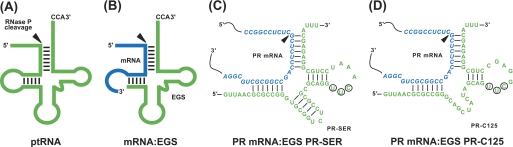
FIGURE 1.
Substrates for RNase P. A, a natural substrate (ptRNA). B, a hybridized complex of a target RNA (e.g. mRNA) and an EGS that resembles the structure of a tRNA. C and D, complexes of PR mRNA sequence with EGS PR-SER and PR-C125. The site of cleavage by RNase P is indicated with an arrowhead. The sequence of the PR mRNA around the targeting site is shown in blue and the EGS sequence is shown in green. The sequences of PR-SER that were equivalent to the T-stem and loop, and variable region of a tRNA molecule were derived from tRNASer, whereas those of PR-C125 were from EGS variant C125. The circled positions in the T-loop of EGS PR-SER and PR-C125 represent the nucleotides that are mutated (5′-UUC-3′ → AAG) to generate PR-SER-C and PR-C125-C, respectively.