Abstract
Topoisomerase II (Top2) activity involves an intermediate in which the topoisomerase is covalently bound to a DNA double-strand break via a 5′-phosphotyrosyl bond. Although these intermediates are normally transient, they can be stabilized by antitumor agents that act as Top2 “poisons,” resulting in the induction of cytotoxic double-strand breaks, and they are implicated in the formation of site-specific translocations that are commonly associated with cancer. Recently, we revealed that TRAF and TNF receptor-associated protein (TTRAP) is a 5′-tyrosyl DNA phosphodiesterase (5′-TDP) that can cleave 5′-phosphotyrosyl bonds, and we denoted this protein tyrosyl DNA phosphodiesterase-2 (TDP2). Here, we have generated TDP2-deleted DT40 cells, and we show that TDP2 is the major if not the only 5′-TDP activity present in vertebrate cells. We also show that TDP2-deleted DT40 cells are highly sensitive to the anticancer Top2 poison, etoposide, but are not hypersensitive to the Top1 poison camptothecin or the DNA-alkyating agent methyl methanesulfonate. These data identify an important mechanism for resistance to Top2-induced chromosome breakage and raise the possibility that TDP2 is a significant factor in cancer development and treatment.
Keywords: Cancer Therapy, DNA Damage, DNA Repair, DNA Topoisomerase, Phosphodiesterases
Introduction
Topoisomerases release torsional stress in DNA and decatenate chromosomes during mitosis (1, 2). A key intermediate of topoisomerase activity is the cleavage complex, within which the topoisomerase cleaves one or both strands of DNA and becomes covalently linked to the 3′ or 5′ terminus of the break via a phosphotyrosyl bond. The cleavage complex is normally transient because the topoisomerase reseals the break at the end of its catalytic cycle. However, cleavage complexes are the targets of a class of antitumor agents that act as topoisomerase “poisons,” prolonging the half-life of the cleaved intermediate and resulting in the formation of potentially cytotoxic DNA strand breaks (3). Whereas topoisomerase I (Top1)4 poisons induce DNA breaks in which the topoisomerase is linked via a phosphotyrosyl bond to the 3′ terminus of the break, topoisomerase II (Top2) poisons induce DNA strand breaks in which the topoisomerase is linked via a phosphotyrosyl bond to the 5′ terminus. Top2-associated DNA breaks account not only for the clinical efficacy of Top2 poisons as antitumor cytotoxic drugs, but are also implicated in genome instability, chromosome translocations, and carcinogenesis (4, 5). Recently, we identified a human 5′-tyrosyl DNA phosphodiesterase (5′-TDP) activity that can cleave 5′-phosphotyrosyl bonds and might thereby release trapped topoisomerase from 5′ termini (6). This enzyme, previously known as TTRAP, is a member of the metal-dependent superfamily of phosphodiesterases and has been implicated in NF-κB signaling (7, 8), ETS1-mediated transcriptional regulation (9), apoptotic regulation (10), and left-right axis determination during development in zebrafish (11). Based on its TDP activity we denoted TTRAP as tyrosyl DNA phosphodiesterase 2 (TDP2) and speculated that this enzyme might play an important role in the repair of Top2-associated double strand breaks (DSBs) induced by Top2 poisons (6). We now demonstrate using avian DT40 cells that TDP2 is the primary if not the only 5′-TDP activity in vertebrate cells and that genetic deletion of TDP2 results in specific and severe sensitivity to Top2 poisons. These data identify TDP2 as a novel cellular component of the DNA repair pathway(s) that protect cells from topoisomerase-induced chromosomal breakage and suggest that TDP2 may be a useful clinical target for inhibition during the chemotherapeutic application of anticancer Top2 poisons.
EXPERIMENTAL PROCEDURES
5′-Tyrosyl DNA Phosphodiesterase Activity
The 19-mer and 20-mer DSB substrates were prepared as described previously (6). Briefly, the 5′-Y 18-bp (5′-YTCCGTTGAAGCCTGCTTT-3′) oligonucleotide was annealed with 19-bp (5′-GAAAGCAGGCTTCAACGGA-3′) or 20-bp (5′-AGAAAGCAGGCTTCAACGGA-3′) oligonucleotides, respectively, and the resulting 1-bp or 2-bp 5′-overhang filled using Klenow DNA polymerase in the presence of [α-32P]dCTP (for the 19-mer) or both [α-32P]dCTP and ddTTP (for the 20-mer). Inclusion of the latter for the 20-mer inhibits degradation of the substrate by nonspecific 3′-nucleases present in whole cell extract. For single-stranded and nicked duplex substrates, the single-stranded 32P-labeled 5′-Y 19-mer prepared as above was gel-purified from denaturing gels and either employed alone (single-stranded substrate) or annealed, along with the 21-bp oligonucleotide (5′-GCGCAGCTAGCGGCGGAT-GGC-3′), to the 40-bp oligonucleotide (5′-GAAAGCAGGCTTCAACGGAGCCATCCGCCGCTAGCTGCGC-3′). Reactions were carried out by incubating the indicated substrate (50 nm) with whole cell protein extract (250 ng/ml) or the indicated concentration of recombinant human His-TDP2/TTRAP protein, which was expressed in Escherichia coli and prepared as described previously (6), in 25 mm HEPES, pH 8.0, 130 mm KCl, 10 mm MgCl2, 1 mm DTT and in the presence or absence of orthovanadate, BSA, or phosphotyrosine-BSA (Sigma) as indicated at 37 °C for 60 min. In reactions containing cell extract 50 μm competitor single-stranded oligonucleotide (5′-CTAACTTGAGCGAAACGGT-3′) was also included to further reduce nonspecific nucleolytic degradation of the duplex substrate. To examine whether the product of hTDP2 activity was a ligatable 5′ terminus, nicked duplex substrate described above was incubated in ligation buffer with hTDP2 and/or human DNA ligase IIIα, which was expressed in E. coli and purified as described previously (12, 13). All reactions were terminated by the addition of formamide loading buffer and fractionated by denaturing PAGE; images were analyzed by phosphorimaging.
Cell Cycle Analysis
Cells grown for the indicated time in the presence or absence of 40 nm etoposide were fixed for 24 h in 70% ethanol at −20 °C. Cells were then washed twice in PBS and 3% BSA, resuspended in PBS containing 10 μg/ml propidium iodide and 100 μg/ml RNase A, and analyzed using a FACS Canto device (Becton Dickinson).
Cell Culture and Transfection
Chicken DT40 B lymphoma cells were cultured in RPMI 1640 medium supplemented with 10−5 m β-mercaptoethanol, penicillin, streptomycin, 10% fetal calf serum (FCS), and 1% chicken serum (Sigma) at 39 °C.
Generation of TDP2 Mutant DT40 Cells
To generate TDP2 gene disruption constructs, TDP2 sequences were amplified by PCR from genomic DNA from DT40 cells (clone 18) using the primer combinations 5′-CCCAAGCTTGGAAAGTACTGTGCTCCGAGT-3′ and 5′-CGCGGATCCGCCCATCAATGTTCCAAGTTAT-3′ for the 5′ arm, and 5′-CGCGGATCCGGATGAGCATGGACAATGAATA-3′ and 5′-CCGCTCGAGGCGCTGTAGGTAGAACAGAAGC-3′ for the 3′ arm. The amplified PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen) and confirmed by sequencing. 2.5-kb HindIII/BamHI (left arm) and 4.4-kb BamHI/XhoI (right arm) DNA fragments were then ligated into pCR2.1-TOPO vector. A single drug-resistance gene (Puro, HisD, or bsr) was then inserted into the BamHI site of the pCR2.1 vector between the TDP2 targeting sequences. To generate TDP2−/−/− cells, 2 × 107 cells were electroporated (Bio-Rad) with 30 μg of the relevant linearized (with XhoI) TDP2 gene-disruption construct and drug-resistant colonies selected by incubation in medium containing 0.5 μg/ml puromycin dihydrochloride (Sigma-Aldrich), 1.0 mg/ml d-histidinol dihydrochloride (Sigma-Aldrich), or 25 μg/ml blasticidine S hydrochloride (Sigma-Aldrich), alone or in combination as appropriate. To detect successful targeting of TDP2 alleles, genomic DNA was isolated from multiple drug-resistant clones, digested with XhoI and EcoRV, and subjected to Southern blot analysis using a 0.8-kb probe (N-probe) amplified using the primers 5′-TGAAGCAAAGCTGACAAGC-TGC-3′ and 5′-CCAGTTAGCGGCAACATTTGAG-3′. Following three consecutive rounds of gene targeting with constructs encoding resistance to Puro, HisD, and Bsr, respectively, two TDP2−/−/− clones (clone 8 and clone 93) were recovered. These clones were employed for phenotypic analysis along with parental DT40 cells and a single TDP2−/−/+ clone (clone H13) in which only two of the three TDP2 alleles were targeted.
Generation of hTDP2 and hTDP2 Catalytic Mutant Transfected DT40 Cells
pcDNA3.1-HisC (Invitrogen) constructs expressing wild-type human TDP2 and TDP2D262A were described previously (note that TDP2 was previously denoted TTRAP) (6). TDP2−/−/− DT40 cells were electroporated with pcDNA3.1-HisC-TDP2, pcDNA3.1-HisC-TDP2 (D262A), or pcDNA3.1-HisC empty vector. After selection in 1.5 mg/ml G418 (Invitrogen) for 6 days, pooled populations of transfectants were collected. For Western blotting, cell extracts were prepared in radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris-Cl, pH 8.0, 1 mm PMSF, and Complete protease inhibitor mixture (Roche)), resolved by SDS-PAGE, and blotted onto nitrocellulose membranes (GE Healthcare). Membranes were blocked for 1 h in PBS-Tween 20 (PBST) containing 5% nonfat dried milk (NFDM) and then incubated for 2 h with affinity-purified anti-TDP2 polyclonal antibody (SY1340) (6) at a 1/200 dilution, in PBST plus 5% nonfat dried milk. Membranes were rinsed in PBST and incubated in PBST plus 5% nonfat dried milk containing horseradish peroxidase-conjugated anti-rabbit IgG (DAKO), as appropriate, at a 1/3,000 dilution for 1 h at room temperature. Membranes were then rinsed with PBST and antibody complexes detected by enhanced chemiluminescence (GE Healthcare).
Clonogenic Survival Assays
To determine sensitivity, cells were plated in 5 ml of medium containing 1.5% (by weight) methylcellulose (Sigma-Aldrich) in 6-well plates at 50, 500, and 5000 cells/well per treatment condition. Media also contained the indicated concentration of methyl methanesulfonate, camptothecin (CPT), or etoposide. In all experiments, cells were incubated for 7–11 days and visible colonies counted. The surviving fraction was calculated by dividing the average number of visible colonies in treated wells by the average number of colonies in untreated wells.
RESULTS
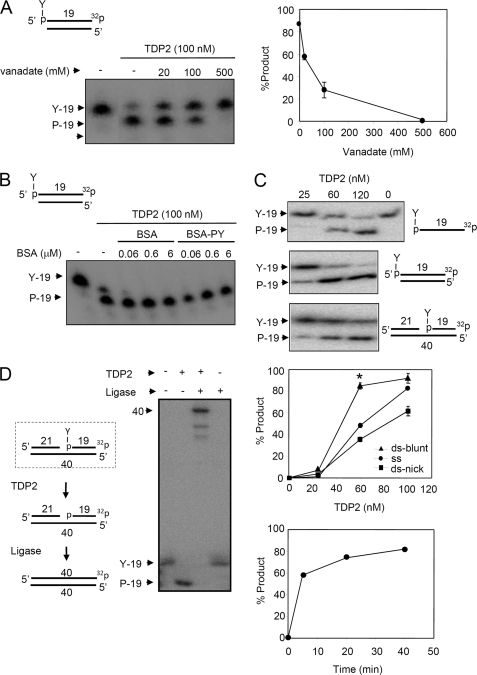
TDP2 (also known as TTRAP) is the first human enzyme with 5′-TDP activity to be identified (6). To characterize this enzyme further we examined its sensitivity to vanadate and its DNA substrate specificity. The 5′-TDP activity of histidine-tagged TDP2 (His-TDP2) on oligonucleotide duplexes harboring 5′-phosphotyrosyl termini was inhibited by vanadate (Fig. 1A). This is similar to the 3′-TDP activity of TDP1 and suggests the involvement of a phosphate transition state during the 5′-TDP reaction. To examine the specificity of His-TDP2 for nucleic acid substrates we first examined whether this activity was inhibited by BSA-tyrosine phosphate, a derivative of BSA containing a phosphotyrosine bond similar to that present in the oligonucleotide 5′-phosphotyrosyl substrate. BSA-tyrosine phosphate failed to inhibit the activity of His-TDP2, even if present at 60-fold higher concentrations than the oligonucleotide 5′-phosphotyrosyl substrate, suggesting that His-TDP2 activity was specific for nucleic acid substrates (Fig. 1B). His-TDP2 was active on a phosphotyrosyl bond present at the 5′ terminus of either a single-stranded oligonucleotide, a DSB, or a single strand break (SSB), but was most active on the DSB substrate (Fig. 1C). This is consistent with a role for TDP2 in the repair of Top2-associated DSBs. However, the activity of His-TDP2 on SSBs with 5′-phosphotyrosyl termini may also be relevant because Top2 also induces this type of break (4). We have suggested that the product of TDP2 activity is a 5′-phosphate terminus that is competent for DNA ligation, based on the sensitivity of the TDP2 product to calf intestinal phosphatase (6). This is an important possibility because if true it would provide an error-free mechanism for repairing topoisomerase-associated DNA breaks by simple DNA ligation. We therefore addressed whether the termini created by TDP2 are ligatable, directly, by employing a substrate that contained a 5′-phosphotyrosyl terminus within the context of a single-stranded nick. Indeed, incubation of this substrate with His-TDP2 and DNA ligase, but not with either His-TDP2 or DNA ligase alone, resulted in conversion of the radiolabeled 19-mer containing the phosphotyrosyl terminus into a radiolabeled 40-mer, confirming that the His-TDP2 generates a ligatable 5′-phosphate terminus (Fig. 1D).
FIGURE 1.
The 5′-TDP activity of His-TDP2 prefers DSBs and liberates ligatable 5′ termini from DNA 5′-phosphotyrosine termini. A, purified recombinant human His-TDP2 (100 nm) was incubated with 50 nm duplex substrate harboring a DSB with a 5′-phosphotyrosine (P-Y) terminus (top) in the absence or presence of the indicated concentrations of orthovanadate for 1 h at 37 °C and reaction products separated by denaturing PAGE and detected by autoradiography. Right, quantification of the fraction (%) of total labeled oligonucleotide converted to 5′-phosphate (19-mer) product is shown. Data are the average (± range) of two independent experiments. B, purified recombinant human His-TDP2 (100 nm) was incubated with duplex substrate as above in the absence or presence of the indicated concentrations of BSA or phosphotyrosine-BSA. C, DNA substrates with a 5′-phosphotyrosine terminus present in single-stranded DNA (top), in a blunt-ended DNA duplex (middle), or in a nicked duplex (bottom), were incubated with the indicated concentrations of His-TDP2 for 1 h at 37 °C and reaction products analyzed as above. The middle graph is the quantification (% of total labeled oligonucleotide converted to 5′-P 19-mer product) of the above experiment, from three independent experiments mean (±S.E.). Asterisk denotes statistically significant difference (p < 0.05; t test) between the indicated blunt-ended DNA duplex (ds-blunt) and single-stranded DNA (ss) data points. The bottom graph is a time course of TDP2 activity (120 nm) on the blunt-ended DNA duplex (50 nm) from a single experiment. D, nicked DNA duplex harboring a 5′-phosphotyrosine terminus (dotted box, left) was incubated with (+) or without (−) 200 nm human His-TDP2 and 50 nm Lig3α and reaction products analyzed as described above. The predicted reaction steps are depicted (left).
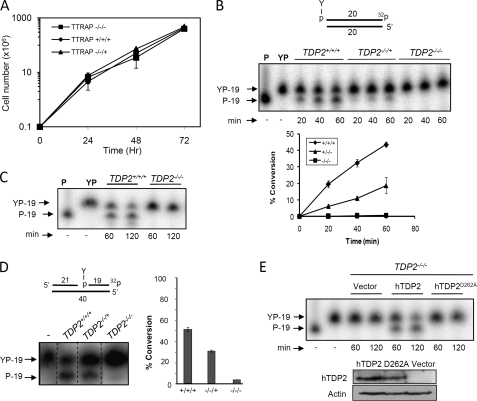
To examine directly the importance of TDP2 for the repair and cellular resistance to Top2-associated DNA breaks we disrupted each of the three TDP2 alleles in chicken DT40 cells, thereby creating a TDP2-deleted mutant cell line (supplemental Fig. 1). The doubling time of TDP2−/−/− cells was not significantly different from that of wild-type DT40 cells, suggesting that cells proliferate at normal rates in the absence of TDP2 (Fig. 2A). TDP2 is currently the only 5′-TDP activity identified in human cells. To address whether other such activities might be present, we compared whole cell extracts from TDP2+/+/+, TDP2−/−/+, and TDP2−/−/− cells for levels of 5′-TDP activity. Whereas whole cell extracts prepared from wild-type TDP2+/+/+ cells readily converted 5′-phosphotyrosyl termini to 5′-phosphate termini at a DSB end, whole cell extracts from TDP2−/−/− cells failed to do so (Fig. 2, B and C). We noted that cell extract from TDP2−/−/+ cells exhibited intermediate levels of 5′-TDP activity, suggesting that a single wild-type TDP2 allele is sufficient to provide detectable levels of 5′-TDP activity (Fig. 2B). Similar observations were observed for 5′-phosphotyrosyl termini present at SSBs (Fig. 2D). Stable expression of wild-type human TDP2 in TDP2−/−/− cells (TDP2−/−/−hTDP2 cells) restored 5′-TDP activity, whereas stable expression of human TDP2D262A in TDP2−/−/− cells (TDP2−/−/− hTDP2 D262A cells) harboring a mutated active site did not, confirming that the absence of 5′-TDP activity in TDP2−/−/− cells was the result of TDP2 deletion (Fig. 2E). Taken together, these data suggest that TDP2 is the major if not the only 5′-TDP activity present in DT40 cell extracts.
FIGURE 2.
5′-Tyrosyl DNA phosphodiesterase activity in TDP2−/−/− cell extracts. A, growth curves of TDP2+/+/+, TDP2−/−/+ (clone H13), and TDP2−/−/− (clone 8) DT40 cells are shown. Data are the mean ± 1 S.D. of three independent experiments. B, duplex substrate harboring a DSB with a 5′-phosphotyrosine terminus (upper) was incubated with whole cell extract from TDP2+/+/+, TDP2−/−/+, and TDP2−/−/− DT40 cells for 20, 40, and 60 min. Lower, the percentage of substrate converted to reaction product is shown (mean ± S.E. from three independent experiments). C, reactions conducted as in B employing TDP2+/+/+ or TDP2−/−/− cell extracts were extended to 120 min. D, duplex substrate harboring a SSB with a 5′-phosphotyrosine terminus (middle) was incubated with whole cell extract from TDP2+/+/+, TDP2−/−/+, and TDP2−/−/− DT40 cells for 20 min. The percentage of substrate converted to reaction product is shown (right) (mean ± S.E. from three independent experiments). E, 5′-TDP activity was analyzed as above in extracts from TDP2−/−/− cells transfected with empty expression vector (Vector) or with expression vector encoding either wild-type human TDP2 (hTDP2) or human TDP2 with a mutated catalytic domain (hTDP2D262A). hTDP2 and actin expression levels were analyzed by Western blotting with anti-hTDP2 and anti-actin antibodies, respectively (bottom). All reaction products were analyzed by denaturing PAGE and phosphorimaging. The positions of substrate (YP-19) and repair product (P-19) are indicated.
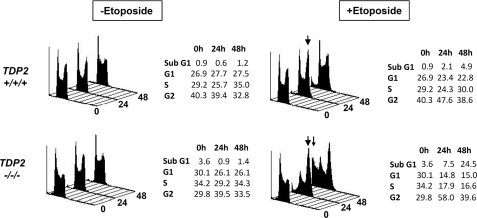
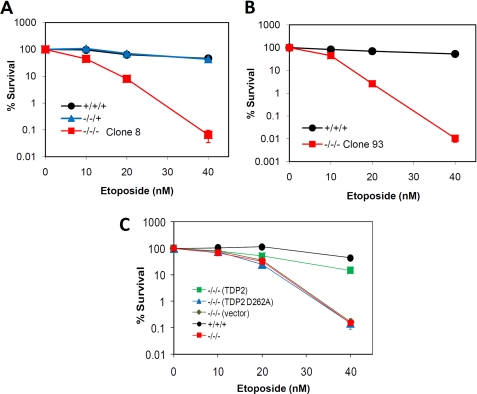
Top2 poisons are an important class of antitumor agents that exert their clinical efficacy by inducing Top2-associated DSBs (14). Based on the 5′-TDP activity of TDP2, we proposed that this enzyme might be important for cellular resistance to Top2 poisons (6). To test this hypothesis, we compared the sensitivity of TDP2+/+/+, TDP2−/−/+, and TDP2−/−/− cells to etoposide, a Top2 poison employed in the treatment of lung, ovarian, and testicular cancer. Flow cytometry revealed that cell cycle progression slowed in both TDP2+/+/+ and TDP2−/−/− cells within 24 h of addition of 40 nm etoposide to the culture medium (Fig. 3). However, whereas the cell cycle profile in TDP2+/+/+ cells returned to near-normal within 48 h, a significant fraction of TDP2−/−/− cells (∼24%) accumulated in a sub-G1 peak, indicative of cell death (Fig. 3, bottom panels). To confirm the hypersensitivity of TDP2−/−/− cells to etoposide we next employed clonogenic survival assays. Strikingly, both of the two independent TDP2−/−/− cell clones generated by gene targeting exhibited high sensitivity to etoposide, with three-to-four log-cell kill observed in TDP2−/−/− cells at etoposide concentrations that were largely nontoxic to TDP2+/+/+ or TDP2−/−/+ cells (Fig. 4, A and B). Importantly, TDP2−/−/− hTDP2 cells expressing human TDP2 exhibited levels of etoposide sensitivity similar to TDP2+/+/+ cells, confirming that the sensitivity of TDP2−/−/− cells was due to loss of TDP2 protein (Fig. 4C). Moreover, TDP2−/−/− hTDP2 D262A cells expressing catalytically impaired or inactive TDP2 were no more resistant to etoposide than were untransfected TDP2−/−/− cells or TDP2−/−/− cells transfected with empty vector, consistent with a requirement for TDP2 phosphodiesterase activity for cellular resistance to etoposide (Fig. 4C). In contrast to etoposide, TDP2−/−/− cells exhibited little or no hypersensitivity to methyl methanesulfonate, a genotoxin that induces DNA base damage that is repaired by the base excision repair pathway (supplemental Fig. 2A). Similarly, TDP2−/−/− cells exhibited little or no hypersensitivity to CPT a Top1 poison that induces DNA strand breaks with Top1-associated DNA 3′ termini (supplemental Fig. 2B). Together, these data demonstrate that TDP2 is a novel component of the DNA damage-response in vertebrate cells and that this enzyme is critical for the survival of these cells to Top2-associated DNA strand breaks.
FIGURE 3.
Accumulation of TDP2−/−/− cells in sub-G1 and G2 following exposure to etoposide. Cell cycle profile of TDP2+/+/+ and TDP2−/−/− (clone 8) DT40 cells that were cultured for 24 or 48 h in the presence or absence of 40 nm etoposide, as indicated, is shown. The accumulation of cells in G2/M or sub-G1 in response to etoposide treatment is indicated by large and small arrowheads, respectively. The fraction of cells with sub-G1, G1, S phase, and G2 DNA content is expressed as a percent of the total cells in each sample and is tabulated.
FIGURE 4.
Cellular hypersensitivity of TDP2−/−/− cells to the Top2 poison, etoposide. A, clonogenic survival of TDP2+/+/+, TDP2−/−/+, and TDP2−/−/− (clone 8) cells cultured in the indicated concentrations of etoposide. B, clonogenic survival of TDP2+/+/+ and TDP2−/−/− (clone 93) cells cultured as above. C, clonogenic survival of TDP2+/+/+ cells, TDP2−/−/− cells (clone 8), and pooled populations of TDP2−/−/− (clone 8) transfectants harboring empty expression vector (vector) or expression vector encoding either wild-type human TDP2 (TDP2) or TDP2 with a mutated catalytic domain (TDP2 D262A). Data are the mean ± S.E. of three independent experiments. Where not visible, error bars are smaller than the symbols.
DISCUSSION
We previously identified human TTRAP as a 5′-tyrosyl DNA phosphodiesterase and consequently denoted this protein TDP2 (6). Here, we have extended the biochemical analysis of TDP2 and addressed, for the first time, the impact of TDP2 deletion on vertebrate cells. The 5′-TDP activity of TDP2 was inhibited by vanadate, suggesting that, like TDP1, cleavage of phosphotyrosyl bonds involves formation of a phosphate transition state. The activity of TDP2 was not inhibited by the presence of excess BSA-tyrosine phosphate, suggesting that TDP activity exhibits specificity for phosphotyrosyl bonds present in nucleic acid substrates. TDP2 exhibited a small preference for 5′-phosphotyrosyl termini at blunt DSBs, compared with single strand 5′ termini or internal 5′ termini at SSBs, consistent with a role of TDP2 in the repair Top2-induced DSBs in vivo. The activity of TDP2 on single stranded termini may be physiologically relevant, however, because the phosphotyrosyl terminus at a Top2 DSB is present as a 4-bp 5′ single stranded overhang. Similarly, the activity of TDP2 on 5′-phosphotyrosyl SSBs may be physiologically relevant because Top2 also induces this type of break, due most likely to incomplete temporal coordination in the cleavage activity of the two Top2 subunits that comprise the active homodimer (4). In addition, topoisomerase III also creates DNA breaks with 5′-phosphotyrosyl termini and preferentially does so in single stranded DNA (15, 16).
To examine the requirement for TDP2 for cellular response to Top2-induced DNA damage we generated an avian DT40 cell line in which all three TDP2 alleles were disrupted. Notably, whereas robust 5′-TDP activity was evident in wild-type cell extracts we detected little or no residual activity in cell extracts from TDP2-deleted cells. This suggests that TDP2 is the primary, if not the only, 5′-TDP activity in higher eukaryotes. In addition, deletion of TDP2 resulted in a high level of hypersensitivity to the Top2 poison, etoposide, and expression of recombinant human TDP2 complemented this hypersensitivity, consistent with TDP2 being a major mechanism for the removal of trapped Top2 from DNA ends. Top2-induced DSBs are composed of 4-bp complementary 5′-overhangs in which the 5′-overhangs harbor the phosphotyrosyl termini. Consequently, because TDP2 can convert 5′-phosphotyrosyl termini into ligatable termini, TDP2 could provide an error-free mechanism for repair of Top2 DSBs in which the only additional DNA processing that is required is DNA ligation. It is likely that TDP2 functions in conjunction with components of the nonhomologous end-joining machinery, with Ku and DNA ligase IV likely controlling events at the DSBs before and/or after TDP2 activity. Consistent with this idea, Lig4−/− and Ku70−/− DT40 cells also exhibit high levels of sensitivity to etoposide (17).
In contrast to etoposide, little or no sensitivity was observed in TDP2−/−/− DT40 cells to methyl methanesulfonate or the Top1 poison CPT. The lack of sensitivity to methyl methanesulfonate is consistent with this agent inducing DNA breaks that possess 5′-sugar phosphate (abasic) termini, rather than 5′-phosphotyrosyl termini. In contrast, CPT induces DNA strand breaks in which Top1 is linked to 3′ termini of DNA breaks via a phosphotyrosyl bond. Although TDP2 possesses weak 3′-TDP activity, it is likely that the primary source of such activity in cells is TDP1, providing a likely explanation for the absence of sensitivity to CPT in TDP2-deleted cells. However, we have shown previously that TDP2 can complement the CPT sensitivity observed in budding yeast cells that lack both Tdp1 and Rad1/Rad10 nuclease-dependent repair mechanisms for 3′-phosphotyrosyl termini (6), raising the possibility that TDP2 may contribute to the survival of vertebrate cells in the absence of TDP1 and/or the Rad1/Rad10 homolog, ERCC1/XPF.
In summary, we describe here the first characterization of a TDP2-deleted cell line and demonstrate that TDP2 is the primary if not the only source of 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cell extracts. Moreover, we show that genetic deletion of TDP2 results in specific and high sensitivity to the Top2 poison and anticancer agent, etoposide, identifying TDP2 as a critical new component of the cellular defense against Top2-induced DNA damage. Top2 damage not only accounts for the clinical efficacy of Top2 poisons, an important and commonly used class of anticancer agents, but also accounts for some of the commonest site-specific translocations observed in human cancer (4, 5). Consequently, TDP2 activity may thus impact significantly both on the development and subsequent treatment of cancer, and inhibitors of this novel enzyme may have clinical utility.
Supplementary Material
This work was supported by Medical Research Council Grants G0600776 and G0901606 (to K. W. C.), Marie Curie IntraEuropean Fellowship 2007-2-1-IEF-221222 (to F. C.-L.), and a Wellcome Trust grant (085284) (to S. F. E. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- Top1
- topoisomerase 1
- Top2
- topoisomerase 2
- CPT
- camptothecin
- DSB
- double strand break
- SSB
- single strand break
- TDP
- tyrosyl DNA phosphodiesterase.
REFERENCES
- 1. Champoux J. J. (2001) Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 2. Wang J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 3. Pourquier P., Pommier Y. (2001) Adv. Cancer Res. 80, 189–216 [DOI] [PubMed] [Google Scholar]
- 4. Deweese J. E., Osheroff N. (2009) Nucleic Acids Res. 37, 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haffner M. C., Aryee M. J., Toubaji A., Esopi D. M., Albadine R., Gurel B., Isaacs W. B., Bova G. S., Liu W., Xu J., Meeker A. K., Netto G., De Marzo A. M., Nelson W. G., Yegnasubramanian S. (2010) Nat. Genet. 42, 668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cortes Ledesma F., El Khamisy S. F., Zuma M. C., Osborn K., Caldecott K. W. (2009) Nature 461, 674–678 [DOI] [PubMed] [Google Scholar]
- 7. Pype S., Declercq W., Ibrahimi A., Michiels C., Van Rietschoten J. G., Dewulf N., de Boer M., Vandenabeele P., Huylebroeck D., Remacle J. E. (2000) J. Biol. Chem. 275, 18586–18593 [DOI] [PubMed] [Google Scholar]
- 8. Rodrigues-Lima F., Josephs M., Katan M., Cassinat B. (2001) Biochem. Biophys. Res. Commun. 285, 1274–1279 [DOI] [PubMed] [Google Scholar]
- 9. Pei H., Yordy J. S., Leng Q., Zhao Q., Watson D. K., Li R. (2003) Oncogene 22, 2699–2709 [DOI] [PubMed] [Google Scholar]
- 10. Zucchelli S., Vilotti S., Calligaris R., Lavina Z. S., Biagioli M., Foti R., De Maso L., Pinto M., Gorza M., Speretta E., Casseler C., Tell G., Del Sal G., Gustincich S. (2009) Cell Death Differ. 16, 428–438 [DOI] [PubMed] [Google Scholar]
- 11. Esguerra C. V., Nelles L., Vermeire L., Ibrahimi A., Crawford A. D., Derua R., Janssens E., Waelkens E., Carmeliet P., Collen D., Huylebroeck D. (2007) Development 134, 4381–4393 [DOI] [PubMed] [Google Scholar]
- 12. El-Khamisy S. F., Saifi G. M., Weinfeld M., Johansson F., Helleday T., Lupski J. R., Caldecott K. W. (2005) Nature 434, 108–113 [DOI] [PubMed] [Google Scholar]
- 13. Taylor R. M., Whitehouse C. J., Caldecott K. W. (2000) Nucleic Acids Res. 28, 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nitiss J. L. (2009) Nat. Rev. Cancer 9, 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson-Sali T., Hsieh T. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7974–7979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson-Sali T., Hsieh T. S. (2002) J. Biol. Chem. 277, 26865–26871 [DOI] [PubMed] [Google Scholar]
- 17. Adachi N., Suzuki H., Iiizumi S., Koyama H. (2003) J. Biol. Chem. 278, 35897–35902 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.