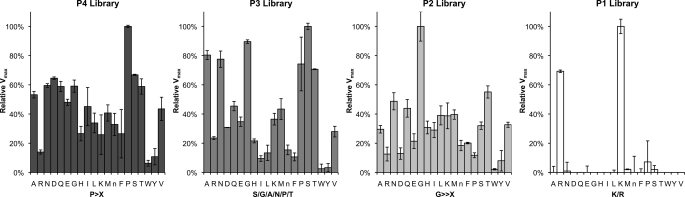
FIGURE 4.
Tetrapeptide substrate specificity profiling of OPB using a positional scanning synthetic combinatorial library. Subsite preference was determined using P1–P4 combinatorial libraries. All of the substrates contained an ACC fluorogenic leaving group. Assays were performed in triplicate, and the mean ± S.D. (error bars) is shown. Activity levels were normalized to the maximum mean value observed for each library. The x axis indicates the amino acid held constant at each position for a given library, designated by the one-letter code (with n representing norleucine). The determined preference for each subsite is in boldface type below the x axis (X indicates that multiple amino acids are permissible in the given subsite).