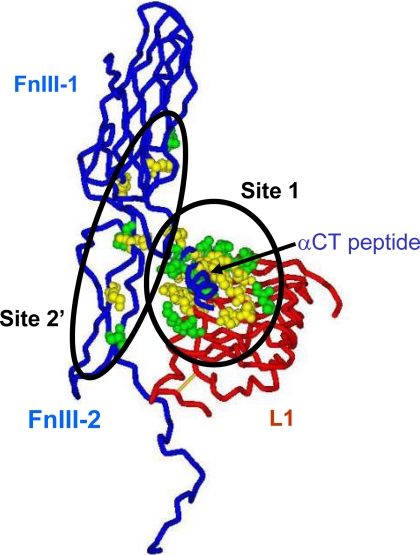
FIGURE 9.
Conservation of receptor residues involved in ligand binding between human and Drosophila insulin receptors. The picture shows the components of one of the two ligand binding sites on the receptor; the L1 domain of one-half human insulin receptor (the backbone in the tube representation is in red), and the FnIII-1 and N-terminal portion of the FnIII-2 domain (interrupted at residue 655 where the insert domain, invisible in the structure (53), starts) as well as the αCT peptide of the second half receptor (backbone in tube representation is in blue). Insulin is thought to cross-link site 1 and site 2′ (shown here as Corey-Pauling-Koltun balls) as mapped by alanine-scanning mutagenesis of the human insulin receptor; see “Discussion” for details. Site 1 is made of a tandem binding element comprising Leu1 residues and the αCT peptide from the opposite receptor half, binding in trans, and site 2 is made of residues at the junction of the FnIII-1 and -2 domains. The residues shown in green are conserved between the human and Drosophila receptors; those shown in yellow are not conserved. Four critical residues from the αCT peptide (from alanine scanning data) also conserved between human and Drosophila receptors are missing from the structure and, therefore, not shown on the figure: Asn711, Val713, Phe714, and Val715. Sequence comparison was made 1) using multiple alignments of the insulin/IGF-1/insulin receptor-related receptors from multiple species genomes in the RILM data base (University College, London, UK) and 2) by aligning the human and Drosophila receptor sequences based on ′blosum62mt2′ score matrix using AlignX software (part of Vector NTI software package; Invitrogen). The two alignments gave identical results for the L1 and FnIII domains, but the αCT peptides are not properly aligned in the RILM data base. PDB accession code: 3LOH. The software used for graphics was DSViewerPro (Accelrys, San Diego, CA).