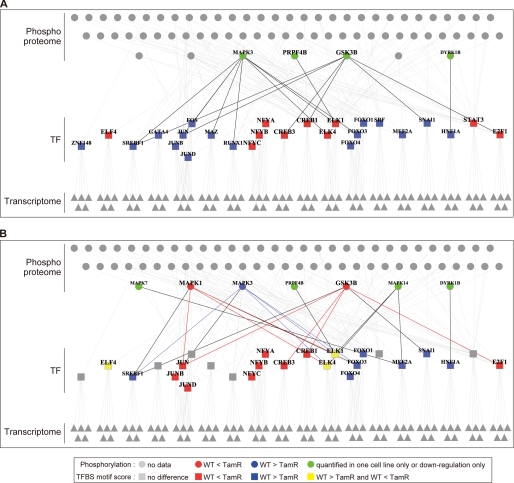
FIGURE 5.
Relationship between transcription factors and their regulatory factors. Transcription factors (TF, rectangles) show different motif activity scores between WT and TamR cells and their regulatory genes (triangles), and signal mediators (circles) are visualized as a network (A, E2; B, HRG). Node color represents phosphorylation and TF activation differences between the two cell lines: red, higher activity in TamR than in WT; blue, higher activity in WT than in TamR; green, phosphorylation change was observed in only one cell or down-regulation only was observed; gray, no change. The edges were color-coded according to TF and regulator activation level in WT and TamR: red, higher activation of TFs and their regulators in TamR than in WT; blue, higher activation of TFs and their regulators in WT than in TamR; black, others. TFs with differential motif activity were inferred from gene expression data (see the supplemental materials for detail), and then factors regulating the predicted TFs were investigated by the KEGG pathway, PhosphoSitePlus, and NetworKIN databases. Relationships extracted from KEGG and PhosphoSitePlus are shown.