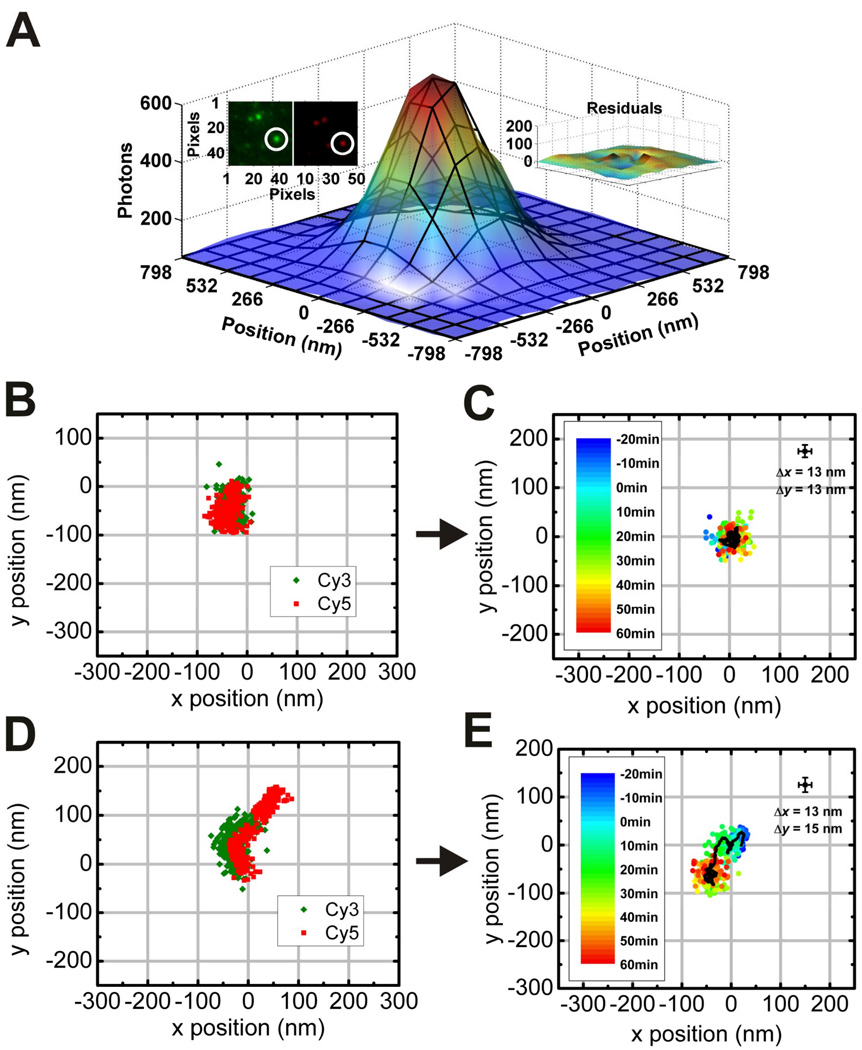
Figure 4.
Extraction of trajectories from a single nanowalker on a linear origami track imaged by TIRF microscopy (Lund et al., 2010). (A) Point spread functions (PSFs) of Cy3-labeled spiders and Cy5-labeled origami are colocalized (inset, upper left) and fit separately to two-dimensional Gaussian functions in each movie frame to determine their coordinates over time. The fit has low residuals (inset, upper right). (B)–(E) Centroids from Gaussian fitting are plotted as a function of time to yield nanowalker trajectories. Even in the absence of the divalent metal ion cofactor, Cy3 and Cy5 coordinates show considerable drift (B), but when the trajectory of Cy5 is subtracted from that of Cy3 it becomes clear that the nanowalker is stationary on its track (C). In contrast, in the presence of 5 mM ZnSO4, subtraction of the raw Cy5 and Cy3 trajectories (D) yields net movement of about 90 nm. In (C) and (E), HBS buffer containing either 0 or 5 mM ZnSO4 was added to the sample at time t = 0 min.