Abstract
A biomimetic nanochannel coated with a self-assembled monolayer (SAM) can be used for sensing and analyzing biomolecules. The interaction between a transported biomolecule and a SAM governs the mechanically or electrically driven motion of the molecule. To investigate the translocation dynamics of a biomolecule, we performed all-atom molecular dynamics simulations on a single-stranded DNA in a solid-state nanochannel coated with a SAM that consists of octane or octanol polymers. Simulation results demonstrate that the interaction between DNA and a hydrophobic or a hydrophilic SAM is effectively repulsive or adhesive, respectively, resulting in different translocation dynamics of DNA. Therefore, with proper designs of SAMs coated on a channel surface, it is possible to control the translocation dynamics of a biomolecule. This work also demonstrates that traditional tribology methods can be deployed to study a biological or bio-mimetic transport process.
Keywords: nanochannel, self-assembled monolayer, DNA, friction
Introduction
Advances in the manufacture of silicon-based nano-devices and the self-assembly of biological or chemical molecules (such as alkanethiol self-assembled monolayers (SAMs)1) have enabled the fabrication of biomimetic devices, which can create biologically compatible environments for sensing and/or analyzing biomolecules. A solid-state nanopore with a SAM-coated surface has been experimentally studied2,3 and could potentially be employed to analyze the transport of ions, water, nutrients or other biomolecules, mimicking a biological protein channel.4,5 When a transported molecule is inside a nanopore, the blockage of an ionic current through the same pore could be used to discern physical features of the molecule. Additionally, the life-time of current blockage, related to the transport speed of the molecule, could be in part affected by the interaction between the molecule and a pore surface. Thus, the selectivity of a biomimetic channel or nanopore could be tuned by adjusting the affinity between a transported molecule and a channel surface, as has been investigated both theoretically6,7,8 and experimentally9. Additionally, to control the motion10 and to sense11,12 a charged biomolecule (such as DNA) in a solid-state nanopore, metal electrodes may be fabricated inside the nanopore whose lifetime can be significantly extended by coating with SAMs to prevent electrochemical reactions, such as corrosion13. While essential to the design of a biomimetic device, the dynamics of a biomolecule through a SAM-coated solid-state nanopore has not been examined. Here, we study, using all-atom molecular dynamics simulations (MD), how a single-stranded DNA (ssDNA) molecule transits solid-state nanochannels coated with designer SAMs with different affinities for ssDNA. The simulations reveal the critical dependence of biomolecular transport through nanochannels on nanotribology.
Simulation Methods
Figure 1 illustrates the simulation setup. Two SAMs are considered: a hydrophilic organosilane octanol and a hydrophobic organosilane octane. The density of coating is about one orgnaosilane molecule per 27 Å2. The silicon atoms in organosilane molecules are positioned in a triangular lattice distorted to lie on the cylindrical surface (radius ~ 2.5 nm), which is shown in the Supporting Information. Other silicon and oxygen atoms in the amorphous solid either fill the space outside or are on the cylindrical surface during melting and quenching processes.14 The average tilt (away from the surface normal) angle of a coated molecule is about 30°. After coating, the channel radius is about 1.3 nm. An ssDNA molecule containing 20 adenine nucleotides is solvated in a 0.1 M KCl electrolyte (explicit water and ions) inside the channel. Periodic boundary conditions are applied in all three dimensions. ssDNA is covalently linked to itself through the periodic boundary (PB). As ssDNA is electrically and hydrodynamically stretched during translocation15, the average spacing (enforced by PB) between neighboring phosphates of the stretched ssDNA in simulation is taken to be 6.8 Å. Water molecules are added to fill each channel (pressure ~ 1 bar); water in the hydrophilic channel is about 30% more than in the hydrophobic one whose surface is covered by a water-vapor layer. The entry of water into a hydrophobic nanochannel might be energetically favorable16 but it can be additionally facilitated by increasing pressure.17 In a previous theoretical study15, ssDNA was harmonically confined to the center of the nanochannel and ions were harmonically pushed away from the channel surface, to prevent ssDNA or ions from being bound to the solid surface. Because of the SAMs, these radial harmonic constraints are released in present simulations. Water and ions are not observed to enter the SAMs. The whole system contains about 56,000 atoms and measures 72×72×136 Å3.
Figure 1.
Simulation system: (a) A top view of an amorphous SiO2 (Si: yellow; O: red) nanochannel containing a layer of octanol coating, a ssDNA molecule and KCl electrolyte. K+ and Cl− ions are shown as tan and cyan spheres. (b) A side and cross-section view. The amorphous SiO2 solid is shown as a molecular surface. For clarity, hydrogen atoms in octanol molecules and ions are not shown. In both (a) and (b), each adenine nucleotide is colored differently and water is not shown. The z-axis is parallel to the channel orientation.
The program NAMD18 was used to perform the all-atom MD simulations reported here using the Bluegene supercomputer. We employed the CHARMM27 force-field19 to model ssDNA and orgnaosilane molecules, and used the TIP3P model to treat the water molecules20. All simulations were carried out in the NVT (T=300 K) ensemble. To maintain the system at constant temperature T, Langevin dynamics was applied to atoms in the SiO2 matrix which were additionally restrained harmonically to occupy amorphous lattice sites.
Results
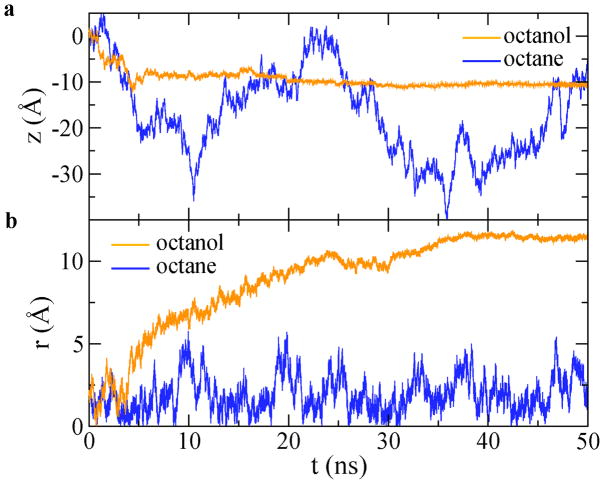
The dependence of the freely diffusive motion of ssDNA on the coating of a solid-state nanochannel is shown in Figure 2. As ssDNA is in a linear conformation, we define the ssDNA position as the center of mass of all phosphorus atoms. The z-component and the radial-component of the ssDNA position are denoted by z and r, respectively. Figure 2a and 2b describe the axial (z) and radial (r) motion of ssDNA, respectively. When ssDNA is in the octane-SAM-coated nanochannel, ssDNA diffuses freely in the axial direction. From the radial motion of ssDNA, the average deviation from the center of the channel is only about 2 Å, i.e. ssDNA is always near the center. When ssDNA is in the octanol-SAM-coated nanochannel, ssDNA moves freely at the beginning. The axial motion fully stops after about 30 ns. Correspondingly, ssDNA moves from the center to the octanol-SAM-coated surface and remains there for the rest of simulation time (also see movies in Supporting Information). This indicates that the contact is effectively “repulsive” between ssDNA and the octane-SAM coating but is effectively “adhesive” between ssDNA and the octanol-SAM coating. These different kinds of contacts therefore can result in different friction on ssDNA during transport.
Figure 2.
The motion of ssDNA in octane-SAM- and octanol-SAM-coated nanochannels, without a driving force on ssDNA. (a) The axial motion of ssDNA. (b) The radial motion of ssDNA.
To quantify the friction force, a key tribological quantity, the steered molecular dynamics method21 was used to mechanically drive ssDNA through the SAM coated channels. Figure 3a illustrates the simulation setup. A spring, whose spring constant is 10 pN/Å, is deployed to drive ssDNA forward, mimicking an optical tweezer pulling of ssDNA through a solid-state nanopore22. One end of the spring is attached to a pulling stage that moves at a constant velocity (v0 = 2 Å/ns), while the other end is attached to the center of mass of all phosphorus atoms. Because of this setup, the pulling force is distributed equally to all phosphorus atoms and as such cannot impart any stretching force to the ssDNA.
Figure 3.
Mechanically driving ssDNA through a SAM-coated nanochannel. (a) simulation setup. v0 is the pulling velocity of the stage (green dot); k is the spring constant; the blue dot represents the ssDNA position. Water and ions are not shown. (b) Force-time dependence when ssDNA is mechanically driven by the harmonic spring in the octanol-SAM-coated nanochannel. (c) Force-time dependence when ssDNA is mechanically driven by the harmonic spring in the octane-SAM-coated nanochannel.
Figure 3b and 3c show time-dependent forces in the spring during a pulling process. Because of the dramatic difference in friction, the dynamics of ssDNA translocation in these two channels are also quite different. After ssDNA diffuses away from the channel center and binds to the octanol-SAM, force in the spring increases linearly with time as shown in Figure 3b. Since the pulling stage moves forward at a constant speed v0, the driving force f in the spring can be written as k(v0t-z). When the force in spring is comparable to the friction force, ssDNA quickly catches up with the pulling stage (a slip event) and the force in spring drops (Figure 3b). When the pulling force is smaller than the friction force, ssDNA is stuck again. As the pulling stage keeps moving forward, force in the spring will build up again and trigger another slip event. So forces in the spring periodically increases and decreases as shown in Figure 3b, corresponding to the stick-slip motion of ssDNA. In the Figure 3b, the value of the first peak (~ 1.9 nN), which can be considered as the static friction force Fs, is bigger than those of following peaks. The kinetic friction force Fk can be estimated from the average pulling force, and is about 1.5 nN. When ssDNA stops, the peak value of friction forces depends on the relaxation time τ for ssDNA to form hydrogen bonds with octanol polymers. If the relaxation time is shorter than the time τ′ for force in the spring to reach the peak value, i.e. τ < τ′ = 2(Fs–Fk)/kv, stick-slip events should be more pronounced. This relation can be satisfied when using a weak spring or a slow pulling velocity. Consequently, subsequent force peaks in Figure 4a should be as high as the first one, if thermally activated processes can be ignored. When τ ≫ τ′, the motion of ssDNA is steady sliding.
Figure 4.
Effects of a SAM coating. (a) Force in the pulling spring f vs. the ssDNA position z. The inset shows the strong interaction between ssDNA (top) and octanol polymers (bottom). (b) Radial motion of ssDNA vs. the ssDNA position.
When ssDNA is in the octane-SAM-coated nanochannel, the average pulling force is much smaller than in the octanol-SAM-coated channel (Figure 3c). As ssDNA is always near the center for the octane-SAM-coated channel during a pulling process, the interaction between ssDNA and the SAM is much weaker. The pulling force in spring is balanced only by the hydrodynamic friction Fh (=ζ v).
The motion of ssDNA described above is further demonstrated in Figure 4, showing the relation between the average driving force < f > and radial motion of ssDNA. When ssDNA is in the octane-SAM-coated nanochannel, the force < f > in the spring is only about 8 pN, as shown in Figure 4a. The radial motion of ssDNA (Figure 4b) shows that ssDNA is not close to the SAM surface. The friction coefficient ζ = < f >/v = 4 pN·ns/Å. When ssDNA is in the octanol-SAM-coated nanochannel, ssDNA gradually moves in the radial direction to the coating surface (Figure 4b). In the beginning, while ssDNA is still near the center, the average driving force in the spring is only a few pico-Newton. But the driving force increases to 1.9 nN after ssDNA is attracted to, and then effectively trapped on, the SAM surface. Note that the measured pulling force (~ 1.9 nN) depends on the pulling velocity (2 Å/ns) that is much faster than what would be used in an actual experiment. The following driven motion of ssDNA is stick-slip like (see below). The inset in Figure 3b illustrates the interaction between ssDNA and octanol polymers. As many as three hydrogen bonds can be formed between a phosphate group in ssDNA and hydroxyl groups in octanol polymers, which explains the found strong friction.
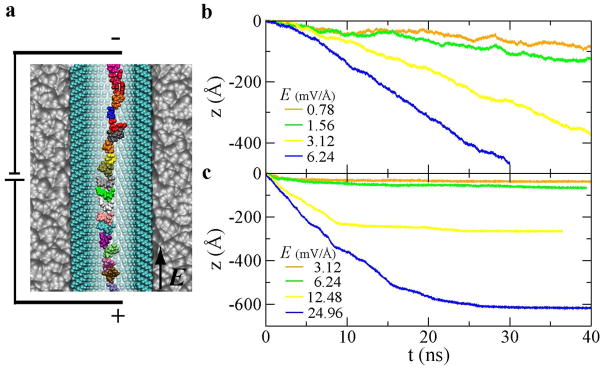
DNA can also be driven through a SAM-coated channel by a biasing electric field as shown in Figure 5a. Figure 5b and 5c show the time-dependent axial motion of ssDNA in the octane-SAM- and octanol-SAM-coated nanochannels, respectively. In the octane-SAM-coated nanochannel, negatively charged ssDNA moves opposite to the field direction. The electric driving force is applied to each negatively charged phosphate group in ssDNA. The slowest speed of simulated electrophoretic motion of DNA is about 2.5 Å/ns, close to the spring pulling velocity v0. The average translocation speed v is linearly proportional to the field strength E. The electrophoretic mobility of ssDNA μ can be computed as, μ = v/E = 2.6 Å2/(ns·mV). Due to the electric screening by counterions and hydrodynamic screening by an electro-osmotic flow on ssDNA surface15, the effective charge of ssDNA qeff =ζμ = 6.5 e (e the charge of an electron) at the ion concentration (0.1 M) examined here. Thus, about 70% of the charge of bare ssDNA (20e) is screened and the effective driving force on ssDNA can be written as f = qeff··E. Note that when ssDNA is in an alpha-hemolysin protein nanopore, even more percentage of the charge of bare ssDNA is screened23.
Figure 5.
Electrically driving ssDNA through a SAM-coated nanochannel. (a) Illustration of the simulation set-up. E is the biasing electric field that drives ssDNA. Water and ions are not shown. (b,c) The electrically-driven axial motion of ssDNA in the octane-SAM- (b) and the octanol-SAM- (c) coated nanochannels
When ssDNA is in the octanol-SAM-coated nanochannel, ssDNA moves in the electric field in the beginning and the axial motion stops after tens of nano-seconds (Figure 5c). Even in a high biasing electric field 24.96 mV/Å, ssDNA can be stuck on the SAM surface of the nanochannel, showing a constant position of ssDNA in Figure 5c. From the radial motion of ssDNA (Figure S2 in Supporting Information), the axial motion stops when ssDNA is close to the SAM surface. During the time when ssDNA moves from the center to the SAM surface, ssDNA moves a longer distance in a higher electric field before ssDNA is stuck on the SAM surface. In applied electric fields, while the thermally activated motion of ssDNA from a stuck state is difficult to study in limited simulation time, a theoretical study of this motion is possible as discussed below.
Discussion
The motion of ssDNA in a SAM-coated nanochannel along the z-axis can be described using a coarse-grain model,
| (1) |
where m is the mass of ssDNA, Fh the hydrodynamic friction force, Fsurf the friction between ssDNA and a SAM surface, f the mechanical or electrical driving force, and FT the instant random force from a thermal bath (such as that used in the Langevin dynamics). In equation 1, Fsurf is unknown and is dependent on properties of a SAM coated inside a nanochannel. In the case of octane-SAM coating, Fsurf is negligible. For ssDNA in the ocatnol-SAM-coated nanochannel, Fsurf, much bigger than Fh, can be modeled by a function varying from −Fs to Fs. Depending on the functional forms of Fsurf and f, the axial motion of ssDNA could be steady-sliding as in the octane-SAM-coated channel or be stick-slip as in the octanol-SAM-coated channel. For example, if the stiffness of a pulling spring is weaker than that of the maximum interfacial stiffness, i.e. k < (dFsurf /dz)max, the motion of ssDNA is stick-slip like assuming that Fsurf (z) is a sinusoidal function24. Additionally, as the electric field driving is the limit of a weak spring pulling24, ssDNA should also advance forward in a stick-slip fashion when electric driving force is comparable to the friction force between ssDNA and a SAM. Note that the theoretical model described in Equation 1 is mathematically similar to the Tomlinson model25 that is deployed to study friction between solid surfaces. Fundamentally, the translocation of ssDNA inside a coated nanochannel is dominated by friction.
Conclusions
It has been demonstrated that the dynamics of ssDNA in a nanochannel is strongly influenced by the tribological effects induced by the choice of SAM coating. In a biasing electric field, the fast ssDNA translocation in an octane-SAM-coated nanochannel results from the weak interaction or low affinity between ssDNA and octane molecules. With the strong affinity between ssDNA and octanol molecules, the translocation of ssDNA is hindered. ssDNA could be stuck on the coating surface intermittently or even permanently. Surface imperfections of a solid-state channel are likely to cause fluctuations in the density and in the surface curvature of a coated SAM, which could affect the interaction between ssDNA and the SAM. These effects will be investigated in the future. The tribological effects stated above can, therefore, be dominant factors influencing the motion of ssDNA. Recently, the importance of interactions between a transported polymer and a protein (alpha-hemolysin) channel has also been demonstrated in experiment.26,27 As it is possible to design and synthesize vast varieties of SAMs,28 the interaction between ssDNA and a SAM can be modified and the translocation speed of ssDNA in a SAM–coated nanochannel could be controlled by the selection of a proper SAM. One important application is to reduce the ssDNA translocation speed, increasing the accuracy for sensing each nucleotide of ssDNA in a solid-state nanopore12. More generally, the study of the transport of a biological molecule in a biomimetic channel, as demonstrated in this work, interfaces both tribology and biology — nanotribolbiology. It therefore is encouraging to apply traditional theories in tribology29 to study biological or bio-mimetic transport processes.
Supplementary Material
Acknowledgments
The authors acknowledge useful discussions with Yuhai Tu and Ajay Royyuru. This work was supported by a grant from National Institutes of Health (R01-HG05110-01).
Footnotes
Supporting Information Available: Arrangement of each molecule in a SAM coated inside a solid-state nanochannel; Radial motion of DNA in an electric field and movies for motion of DNA in SAM-coated nanochannels. This Material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Holmlin R, Haag R, Chabinyc M, Ismagilov R, Cohen A, Terfort A, Rampi M, Whitesides G. Electron Transport through Thin Organic Films in Metal-Insulator-Metal Junctions Based on Self-Assembled Monolayers. J Am Chem Soc. 2001;123:5075–5085. doi: 10.1021/ja004055c. [DOI] [PubMed] [Google Scholar]
- 2.Wanunu M, Meller A. Chemically Modified Solid-State Nanopores. Nano Lett. 2007;7:1580–1585. doi: 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- 3.Yameen B, Ali M, Neumann R, Ensinger W, Knoll W, Azzaroni O. Synthetic Proton-Gated Ion Channels via Single Solid-State Nanochannels Modified with Responsive Polymer Brushes. Nano Lett. 2009;9:2788–2793. doi: 10.1021/nl901403u. [DOI] [PubMed] [Google Scholar]
- 4.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasianowicz JJ, Robertson JW, Chan ER, Reiner JE, Stanford VM. Nanoscopic porous sensors. Annu Rev Anal Chem. 2008;1:737–766. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 6.Berezhkovskii A, Bezrukov S. Optimizing Transport of Metabolites through Large Channels: Molecular Sieves with and without Binding. Biophys Lett. 2005;88(3):L17–L19. doi: 10.1529/biophysj.104.057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong CTA, Muthukumar M. Polymer translocation through α-hemolysin pore with tunable polymer-pore electrostatic interaction. J chem Phys. 2010;133:045101–045112. doi: 10.1063/1.3464333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lubensky DK, Nelson DR. Driven Polymer Translocation through a Narrow Pore. Biophys J. 1999;77(4):1824–1838. doi: 10.1016/S0006-3495(99)77027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iqbal S, Akin D, Bashir R. Solid-state nanopore channels with DNA selectivity. Nat Nanotech. 2007;2:243–248. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 10.Polonsky S, Rossnagel S, Stolovitzky G. Nanopore in metal-dielectric sandwich for DNA position control. Appl Phys Lett. 2007;91:153103–153105. [Google Scholar]
- 11.Tsutsui M, Taniguchi M, Yokota K, Kawai T. Identifying single nucleotides by tunnelling current. Nat Nanotech. 2010;5:286–290. doi: 10.1038/nnano.2010.42. [DOI] [PubMed] [Google Scholar]
- 12.Branton D, Deamer D, Marziali A, Bayley H, Benner S, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich S, Krstic P, Lindsay S, Ling X, Mastrangelo C, Meller A, Oliver J, Pershin Y, Ramsey J, Riehn R, Soni G, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss J. The potential and challenges of nanopore sequencing. Nat Biotech. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrer S, Ahmed S, Afzali-Ardakani A, Luan B, Waggoner P, Shao X, Peng H, Goldfarb D, Martyna G, Rossnagel S, Deligianni H, Stolovitzky G. Electrochemical Characterization of Thin Film Electrodes Towards Implementing a DNA-Transistor. Langmuir. doi: 10.1021/la102671g. (in press) [DOI] [PubMed] [Google Scholar]
- 14.Cruz-Chu E, Aksimentiev A, Schulten K. Water-silica force field for simulating nanodevices. J Phys Chem B. 2006;110:21497–21508. doi: 10.1021/jp063896o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan B, Aksimentiev A. Electro-osmotic screening of the DNA charge in a nanopore. Phys Rev E. 2008;78:021912–021915. doi: 10.1103/PhysRevE.78.021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummer G, Rasaiah J, Noworyta J. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 17.Striolo A, Chialvo A, Gubbins K, Cummings P. Water in carbon nanotubes: Adsorption isotherms and thermodynamic properties from molecular simulation. J Chem Phys. 2005;122(23):234712. doi: 10.1063/1.1924697. [DOI] [PubMed] [Google Scholar]
- 18.Phillips J, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel R, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 20.Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 21.Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr Opin Struct Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 22.Keyser U, Koeleman B, Dorp S, Krapf D, Smeets R, Lemay S, Dekker N, Dekker C. Direct force measurements on DNA in a solid-state nanopore. Nat Phys. 2006;2:473–477. [Google Scholar]
- 23.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA transport into an asymmetric nanometer-scale pore. Phys Rev Letts. 2000;8:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 24.Luan B, Peng H, Polonsky S, Rossnagel S, Stolovitzky G, Martyna G. Base-By-Base Ratcheting of Single Stranded DNA through a Solid-State Nanopore. Phys Rev Lett. 2010;104:238103–238106. doi: 10.1103/PhysRevLett.104.238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomlinson G. A molecular theory of friction. Phil Mag Series. 1929;7:905–939. [Google Scholar]
- 26.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JWF. Theory for polymer analysis using nanopore-based single-molecule mass spectrometry. Proc Natl Acad Sci. 2010;107:12080–12085. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson JWF, Rodrigues CG, Stanford VM, Rubinson KA, Krasilnikov OV, Kasianowicz JJ. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc Natl Acad Sci. 2007;104:8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulman A. Formation and Structure of Self-Assembled Monolayers. Chem Rev. 1996;96:1533–1554. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 29.Nosonovsky M, Bhushan B. Multiscale friction mechanisms and hierarchical surfaces in nano- and bio-tribology. Mater Sci Eng. 2007;58:162–193. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.