Abstract
The procedure of drug approval is time-consuming, costly and risky. Accidental findings regarding multi-specificity of approved drugs led to block-busters in new indication areas. Therefore, the interest in systematically elucidating new areas of application for known drugs is rising. Furthermore, the knowledge, understanding and prediction of so-called off-target effects allow a rational approach to the understanding of side-effects. With PROMISCUOUS we provide an exhaustive set of drugs (25 000), including withdrawn or experimental drugs, annotated with drug–protein and protein–protein relationships (21 500/104 000) compiled from public resources via text and data mining including manual curation. Measures of structural similarity for drugs as well as known side-effects can be easily connected to protein–protein interactions to establish and analyse networks responsible for multi-pharmacology. This network-based approach can provide a starting point for drug-repositioning. PROMISCUOUS is publicly available at http://bioinformatics.charite.de/promiscuous.
INTRODUCTION
Quality plays a crucial role in the highly competitive drug development process. However, cost and the time invested also affect decisions and push scientists to develop new technologies and methods.
In the past few decades the de novo development of drugs has become more and more challenging. About 90% of drugs fail during development in phase 1 clinical trials, which makes this process extremely expensive and time consuming (1). To bring a single de novo drug to the market, an average of more than $800 million is spent in a time period of 15 years; with costs varying from $500 to $2000 million depending on the developing company or the therapy (2). Hence the number of new drugs introduced to the market has not kept in line with the cost of research and development. The cost of a failure is higher by orders of magnitude in the latter stages of development (3). Therefore, effective and innovative approaches are required in the development process. Addressing the problem of off-targets during the design phase, will lead to faster and more efficient drug-development which will make it possible to save patients’ lives and alleviate their suffering. In addition, much effort in terms of financial risk and time could be saved.
The most important causes of drug failures are a lack of efficacy and toxicity (4–6). Limited drug efficacy can be caused by the intrinsic robustness of the biological network of which the intended target is a part (7); whereas toxicity of a drug may be caused by unwanted cross-reactivity with other biologically relevant targets (8). Furthermore, the intended drug target might exhibit previously unknown functions in other processes within the cell or in other tissues.
These issues call for innovative approaches reflecting the insights that no target stands alone, but is embedded in a highly complex and heterogeneous network. There is nothing like a one-to-one relation between a drug and its target; cross reactivity of different strengths with other targets must be considered.
Unfortunately, the development of new drugs according to these insights is hampered by the growing but still limited knowledge about biological networks. Since systems biology attempts to extend the knowledge of these networks, its combination with drug development promises huge advantages to both fields (9,10).
A good example of the knowledge transfer between drug development and systems biology was presented by Campillos et al. (11). In this work it was shown that known side-effects and structural similarity of drugs can successfully be used to identify new targets for known drugs. This method has been applied successfully, for example leading to a patented reposition of Aprepitant (12). In another case, Kinnings et al. were able to reposition a safe drug as a promising lead compound for a new class of anti-tubercular therapeutics using off-target information (13).
The most prominent example of successful drug repositioning was Sildenafil which was initially studied for use in hypertensia and angina pectoris, but has been repositioned as a treatment for erectile dysfunction, and is now known by the trade name of Viagra (14).
As is the case with the aforementioned drug, until now most cases of drug repositioning are the result of serendipitous observations. One of the few successful systematic approaches was performed by Keiser et al. (15) where a network of drug–target connections was constructed by representing target similarity in terms of respective ligand structural similarities. This knowledge was used to construct a network of predicted drug–target connections implying novel targets for known drugs. Another example was that of Xie et al. (16) in explaining the off-target effects of the CETP inhibitor Torcetrapib that was taken out of phase III clinical trials after it was discovered to have significant side-effects. Their work revealed a complex network of interactions with up to twelve putative off-targets.
Plenty of data on this topic are publicly available, often free for academic users. However, it is scattered over different resources, which only have a small overlap. Therefore, a comprehensive analysis of the available data was not possible until now. PROMISCUOUS is an exhaustive network-focused resource of protein–protein and protein–drug interactions enriched with side-effects and structural information which aims to provide a uniform data set for further analysis, integrating basic graph theoretical analysis methods. This resource forms a unique starting point for indication finding and drug-repositioning. Furthermore, it enables the exploration and understanding of off-target-effects and the general analysis of the interplay between drugs and targets.
DATABASE
Integrated data
PROMISCUOUS contains three different types of entities: drugs, proteins and side-effects. Proteins are retrieved from UniProt (17) and displayed with synonyms and organism information. If available, 3D-structures from PDB (18) and EC-numbers (19) are given. About 25 000 drugs from SuperDrug (20) were integrated into the database and assigned metadata as ATC-codes (WHO-classification: Anatomical Therapeutic Chemical) (21), structure information and synonyms. A total of 1100 different side-effects related to the drugs contained in PROMISCUOUS were collected from SIDER (22) and integrated into the dataset.
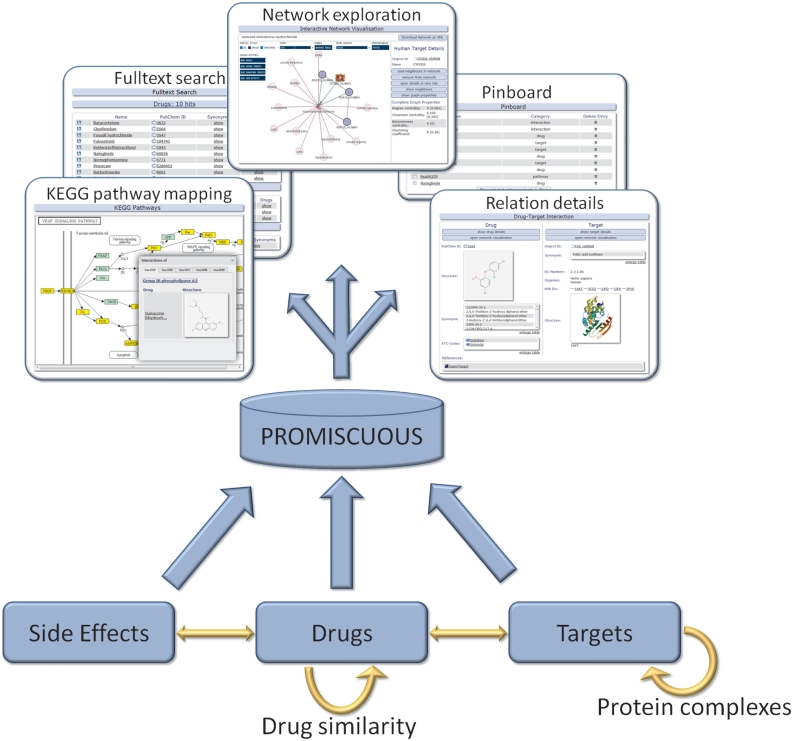
The function of a drug or protein (target) in an organism results from its interactions with other entities. PROMISCUOUS integrates relations between drugs, targets and side-effects as depicted schematically in Figure 1. The entities contained in PROMISCUOUS are connected to each other through drug–target, drug side-effect, protein–protein and drug–drug relations.
Figure 1.
Schematic representation of PROMISCUOUS: the database integrates three types of entities: drugs, targets and side-effects. These are connected by different types of relationships.
To provide a comprehensive dataset of drug–target relations, PROMISCUOUS integrates drug–target relations from the extensive databases DrugBank (23), SuperTarget (24) and SuperCyp (25), as well as newly explored ones as follows. Complete MEDLINE/PubMed data were downloaded from the NCBI FTP site in XML format. The LingPipe-package (26) was used to parse the MEDLINE data from its native XML format into structured Java objects. To create a text-index, objects were transferred to the Lucene package, provided by the Apache Software Foundation (27). Both tools are free open-source software implemented in Java. Two lists were used for the searches containing the (i) drug- and (ii) target-synonyms. The indexed data fields were then searched by dynamically combining the two lists with the query language provided by Lucene and LingPipe. For completeness, keywords were searched in the abstracts, titles and MeSH (Medical Subject Headline) fields. NER-algorithms (named entity recognition) were applied to identify drug- and target-entities in the indexed PubMed abstracts. These entities were linked to each other by rule-based algorithms and sorted by their relevance for drug–target relations, e.g. by the distance between entities. In a further step, the resulting 6300 papers with the highest rank were manually validated to identify false positives. Therefore, a validation website was set up where curators were enabled to login and validate the presumed drug–target relations in the literature. To put the entities into a cellular context the drugs and targets contained in the database were mapped onto 1600 KEGG (28) pathways.
Knowledge about the way target-proteins interact with each other is necessary in order to optimize drug-action. A dataset that integrates physical protein–protein interactions from several databases was retrieved from ConsensusPathDB (29) and integrated into PROMISCUOUS.
Drugs stored in PROMISCUOUS assume inferred relationships through structural similarity. A server for drug–target prediction [SuperPred (30)] was formerly developed in our group and is now used by PROMISCUOUS to provide the user with drugs which have a high probability of acting in a similar way on a target.
The different entities in PROMISCUOUS are interconnected by the relation types described above. They form a large network consisting of 12 000 proteins and 104 000 associated interactions, as well as 21 500 relations connecting 5000 drugs with 6500 target-proteins, often annotated with PubMed-IDs. This information is enriched by 63 000 side-effect–drug associations and information on protein complex composition (31). The database will be continuously maintained and annually updated.
Exploration by user queries
The data delivered by PROMISCUOUS can be queried through various search forms. The easiest way to search for a drug is by its name or PubChem ID. Alternatively, given that drugs in PROMISCUOUS are classified by ATC-code makes it possible to find drugs with a specific desired medical indication. As stated, knowledge on side-effects can be extremely useful in drug-repositioning; therefore searching drugs by their side-effects has also been implemented in PROMISCUOUS. Targets can be queried by the protein name and different commonly used identifiers such as Uniprot ID, Accession number, PDB ID or KEGG ID. Where proteins were assigned EC-numbers, these also can be used as search terms.
PROMISCUOUS can also be explored by metabolic and signalling pathways. These are retrieved from KEGG via web service. Targets for which drug–target relationships are available are highlighted; information on drugs addressing these relationships are displayed by hovering the mouse pointer above them. Information on pathway affiliation is available for every drug or target. To facilitate the exploration of PROMISCUOUS a ‘pin board’ was implemented. It enables the user to store drugs, targets and pathways of interest in order to save the search and repeat it later in the same session. Alternatively, the user can choose one or more objects stored in the ‘pin board’ and load them into the network visualization as discussed below.
Network-based exploration of the data space
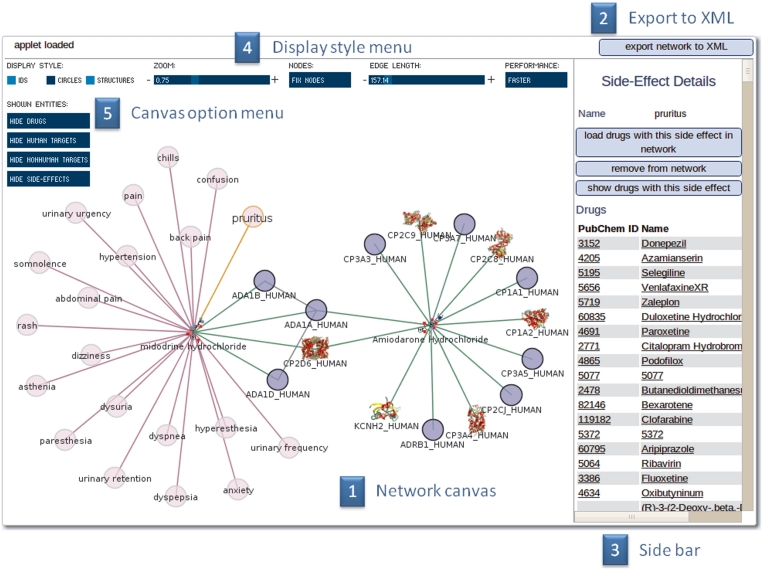
For a scientific yet intuitive way of exploring and handling the data a Java plug-in for interactive network visualization was developed. The interface represents database entities (drugs, targets and side-effects) as nodes in a network with edges representing the relations between them. Starting from an arbitrary set of drugs and targets, this network can be explored interactively. By double clicking a node neighbouring nodes are loaded into the network in real time. That allows users to construct a personalized complex network of drug–target side-effects data. Furthermore, detailed information about nodes and edges, as well as additional features are available via the bar on the right side of the applet (Figure 2). For example basic graph properties (Betweeness, Degree, Clustering Coefficient) for the proteins in the display and the complete protein–protein-interaction-network can be calculated. It is possible to save the user-defined network as an XML file to the local client at any time and to load this XML representation into the interface again later on.
Figure 2.
Network visualization interface. (1) The main window enables the user to view and manipulate the drug–target side-effects network. By double clicking a node neighbour nodes are loaded into the current network. (2) The current state of the network can be saved at any time into an XML file. (3) The side bar shows context sensitive information about highlighted nodes, outlinks to more detailed information tabs, groups of highlighted nodes and graph/node properties. (4) To establish a clear view of the network the user is able to manipulate the display style of the nodes, to fix the view, zoom in and out and to alter the spring constant of the edges. (5) The nodes can be represented by different drawing styles. If a PDB structure of a target is available, the target is an icon like representation of the crystal structure. The nodes can be hidden and redrawn.
EXAMPLES OF USE
The following two use cases illustrate the various features of PROMISCUOUS.
Case study 1: Memantine and Amantadine: use of the PROMISCUOUS drug similarity feature
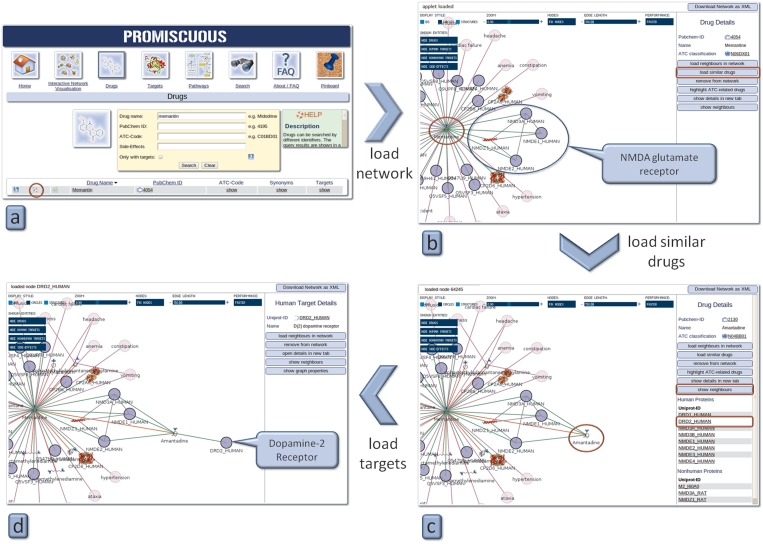
Memantine is a drug prescribed for dementia in patients with Alzheimer’s disease. It acts as a non-competitive antagonist for N-methyl-d-aspartate (NMDA) glutamate receptors. One of its known side-effects is vomiting; in this case study a possible explanation is sought for this side-effect. Starting with Memantine as a drug search, one hit is found (Figure 3a). By opening the interactive network visualization tool, Memantine is shown with a subset of its targets and side-effects, among them the NMDA glutamate receptor and vomiting as a side-effect (Figure 3b). Next, similar drugs to Memantine are loaded into the network by first clicking on the drug and then on the button labelled ‘show similar drugs.’ One of them, the anti-Parkinson drug Amantadine, shares the NMDA glutamate receptor as a target with Memantine (Figure 3c). Besides this receptor, Amantadine targets the Dopamine-2-Receptor, known to be linked to vomiting (click on Amantadine and then on the button labelled ‘show neighbours’) (Figure 3d). Based on the fact that similar drugs often act on the same targets, one can assume that Memantine may also act on the Dopamine-2-Receptor, thereby causing vomiting as a side-effect.
Figure 3.
Case study 1—use of the PROMISCUOUS drug similarity feature. The red circles depict where to click in order to reproduce the example.
Case study 2: Mirtazapine: use of the PROMISCUOUS side-effect similarity feature
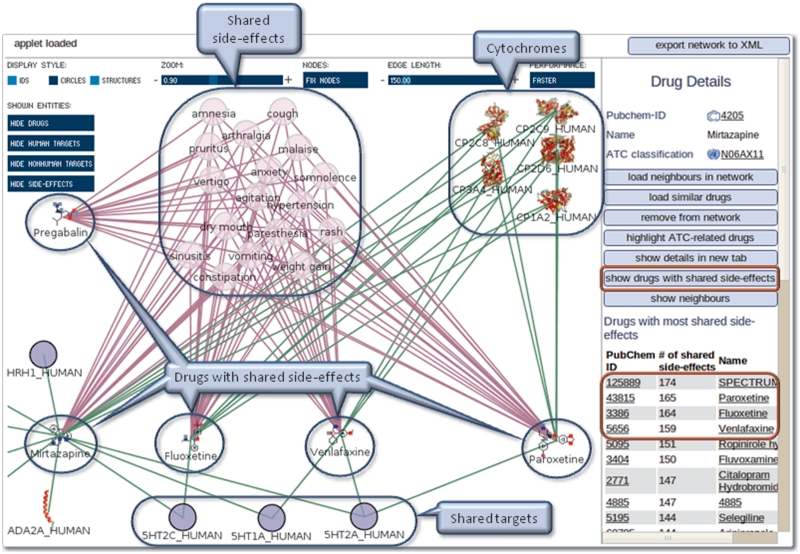
Mirtazapine is an antidepressant for which 184 side-effects are detailed in PROMISCUOUS. Based on this side-effect information, a search for related drugs can be performed in the network exploration tool. After selecting a drug, for example Mirtazapine, a list of other drugs sharing side-effects with it can be retrieved by clicking on ‘show drugs with shared side-effects’ (Figure 4). The list is sorted based on the number of shared side-effects. The drugs with the highest numbers of shared side-effects are expected to act on similar targets. Loading the four drugs with the most shared side-effects (Fluoxetine, Venlafaxine, Paroxetine, Pregabalin) indeed shows that three of them not only share side-effects but also their targets (serotonin receptors) and cytochromes with Mirtazapine. This demonstrates that this feature is able to find drugs acting on similar targets based on similar side-effect information.
Figure 4.
Case study 2—use of PROMISCUOUS side-effect similarity feature. Not all targets and side-effects are shown for clarity. The red circles depict where to click in order to reproduce the example.
CONCLUSIONS AND FUTURE DIRECTIONS
With PROMISCUOUS we developed a rich source of information about drug and target related interactions. The database not only contains drug–target interaction data, but also protein–protein interaction and drug side-effect data, which have proven to be useful for drug repositioning. This information is mapped to its biological context via KEGG pathways. The integrated network visualization tool allows the network of the intended target to be explored in an intuitive way. Additionally, the implemented features like showing common neighbours, computing graph properties, selecting drugs with the same ATC-codes and loading similar drugs allow interactive analyses to be performed on the network. These analyses have proven to be useful in order to identify candidates for drug repositioning. However, due to the complex nature of drug–target interactions these candidates have to be validated and tested experimentally.
Currently, the constructed network can be saved as a proprietary XML file. Soon, supporting of common file formats such as XGMML or SBML is planned to be implemented in order to be compatible with other network visualization software like Cytoscape (32).
PROMISCUOUS is publicly available without registration; it is licensed under a Creative Commons Attribution- Non-commercial-Share Alike 3.0 License.
FUNDING
International Research Training Group on Genomics and Systems Biology of Molecular Networks (GRK1360); German Federal Ministry of Education and Research (MedSys); European Commission (SynSys). Funding for open access charge: MedSys, SynSys, Deutsche Forschungsgemeinschaft SFB 449.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Robert Adams for his stimulating input and Catherine Sargent for her advice as well as Daniel Pommer, Robert Lehmann, Arno Wuithschick, Christian Hoppe and Stefan Günther for their help in the text mining-process.
REFERENCES
- 1.Krantz A. Diversification of the drug discovery process. Nat. Biotechnol. 1998;16:1998–1998. doi: 10.1038/4243. [DOI] [PubMed] [Google Scholar]
- 2.Adams CP, Brantner VV. Estimating the cost of new drug development: is it really 802 million dollars? Health Affairs (Project Hope) 2006;25:420–428. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- 3.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 4.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 5.Kola I. The state of innovation in drug development. Clin. Pharmacol. Therap. 2008;83:227–230. doi: 10.1038/sj.clpt.6100479. [DOI] [PubMed] [Google Scholar]
- 6.Pearson H. The bitterest pill. Nature. 2006;444:532–523. doi: 10.1038/444532a. [DOI] [PubMed] [Google Scholar]
- 7.Kitano H. A robustness-based approach to systems-oriented drug design. Nat. Rev. Drug Discov. 2007;6:202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- 8.Weber A, Casini A, Heine A, Kuhn D, Supuran CT, Scozzafava A, Klebe G. Unexpected nanomolar inhibition of carbonic anhydrase by COX-2-selective celecoxib: new pharmacological opportunities due to related binding site recognition. J. Med. Chem. 2004;47:550–557. doi: 10.1021/jm030912m. [DOI] [PubMed] [Google Scholar]
- 9.Butcher EC, Berg EL, Kunkel EJ. Systems biology in drug discovery. Nat. Biotechnol. 2004;22:1253–1259. doi: 10.1038/nbt1017. [DOI] [PubMed] [Google Scholar]
- 10.Lum PY, Derry JM, Schadt EE. Integrative genomics and drug development. Pharmacogenomics. 2009;10:203–212. doi: 10.2217/14622416.10.2.203. [DOI] [PubMed] [Google Scholar]
- 11.Campillos M, Kuhn M, Gavin A, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn M, Campillos M, Bork P, Jensen LJ, Gavin A, Costi MP, Luciani R, Preissner R, Fan H, Hossbach J, et al. Use of Aprepitant and Dericatives Thereof for the Treatment of Cancer. 2008. Patent WO 2009/124756 A1; PCT/EP2009/002621. [Google Scholar]
- 13.Kinnings SL, Liu N, Buchmeier N, Tonge PJ, Xie L, Bourne PE. Drug discovery using chemical systems biology: repositioning the safe medicine comtan to treat multi- drug and extensively drug resistant tuberculosis. PLoS Comput. Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booth B, Zemmel R. Quest for the best. Nat. Rev. Drug Discov. 2003;2:838–841. doi: 10.1038/nrd1203. [DOI] [PubMed] [Google Scholar]
- 15.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie L, Li J, Xie L, Bourne PE. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput. Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Uniprot Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Bhat TN, Gilliland G, Shindyalov IN, Weissig H, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goede A, Dunkel M, Mester N, Frommel C, Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 21.WHO Expert Commitee. World Health Organization Technical Report Series. 2008. The selection and use of essential medicines. (950) p. backcover, vii-174. [PubMed] [Google Scholar]
- 22.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol. Sys. Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Günther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, Guenther S, Winnenburg R, Schroeder M, Preissner R. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2009;38:D237–D243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alias-i (2008) Alias-i. Lingpipe 3.8.2. http://alias-i.com/lingpipe/index.html (May 2010, date last accessed)
- 27.McCandless M, Hatcher E, Gospodnetić O. Lucene in Action. USA: Manning Publications; 2010. [Google Scholar]
- 28.Kanehisa A, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunkel M, Günther S, Ahmed J, Wittig B, Preissner R. SuperPred: drug classification and target prediction. Nucleic Acids Res. 2008;36:W55–W59. doi: 10.1093/nar/gkn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]