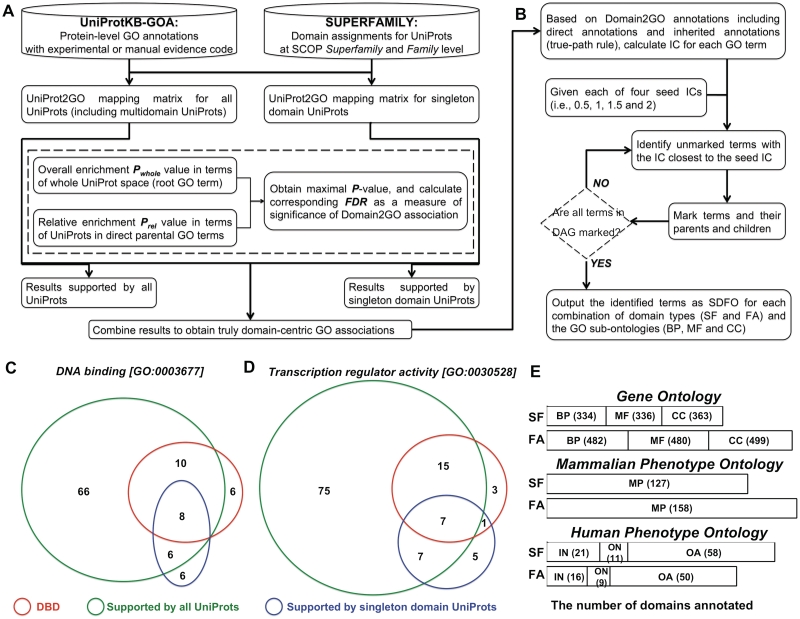
Figure 2.
Functional and phenotypic annotations of structural domains at the SCOP superfamily (SF) and family (FA) levels. (A) Flowchart of inferring domain-centric GOAs using UniprotKB-GOA database and domain assignments in SUPERFAMILY database. (B) Illustration of the procedure to create SDFO based on information theoretic analysis of Domain2 GOA profiles. (C) Venn diagram in which the area of each region is proportional to the differences and intersections among domains annotated to a GO term `DNA binding’ [GO:0003677] using all UniProt sequences (90, circled in green), domains annotated to the term only using singleton domain UniProt sequences (20, circled in blue), and domains in DBD which can be found in at least one UniProt sequence annotated to the term (24, circled in red). (D) Venn diagram showing the differences and intersections among domains annotated to a GO term `transcription regulator activity’ [GO:0030528] using all UniProt sequences, only using singleton domain UniProt sequences, and in DBD which can be found in at least one UniProt sequence annotated to the term. (E) The total number (shown in parenthood) of domains annotated to ontologies. GO depicts three biological concepts: BP, Biological Process; MF, Molecular Function; CC, Cellular Component. Results are based on Domain2 GOAs supported both by singleton domain UniProt sequences and all UniProt sequences. In MPO, it describes mammalian phenotype (MP) related to the mouse with a specific genetic mutation. HPO has three sub-ontologies: IN, inheritance; ON, onset and clinical course; OA, organ abnormality.