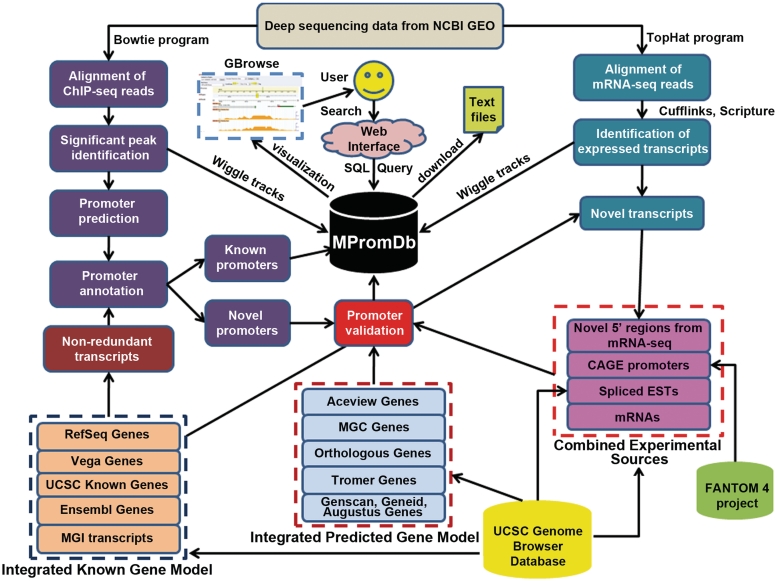
Figure 1.
The block diagram and workflow of updated MPromDb database. Deep sequencing datasets were downloaded from NCBI GEO server and processed by our analysis and annotation pipeline. The identified promoters are deposited in MPromDb tables. Novel promoters are compared to various existing experimental and predicted gene promoter regions and status of novel promoters is deposited in the relational tables. The database is accessed through a user-friendly webpage. The database is integrated with open source genome browser (GBrowse) to visualize the promoter and various ChIP-seq enrichment profiles.