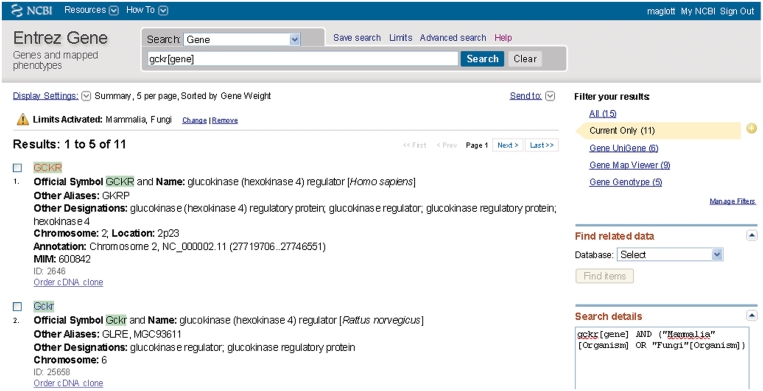
Figure 1.
Representative ‘Summary’ report of query results. Result (partial) of a query to retrieve information about gckr as a gene symbol in mammals or fungi. This figure illustrates several points: (i) use of field restriction in the query; (ii) the display when ‘Limits’ is invoked to restrict results, in this case by species; (iii) use of ‘Display Settings’ to report five records per page ordered by Gene Weight (computed by number of gene-specific citations and conservation) and (iv) use of MyNCBI to highlight matches to the query term in the result set in green. ‘Limits Activated’: Mammalia, Fungi indicates that Mammalia and Fungi were both checked on the form accessed from ‘Limits’ over the query bar. Of the 15 results that were returned, the information under ‘Filter your results’ at the upper right indicate that 11 are current (Current Only, highlighted), 5 have genotype information available in dbSNP (Gene Genotype), 9 can be viewed in Map Viewer (Gene Map Viewer) and 8 have expression data in UniGene (Gene UniGene). For each GeneID returned by the filtered query, the summary includes the species, preferred and alternate symbols, preferred and other descriptive names, chromosome localization, the GeneID and the MIM number when appropriate. Click on any symbol to link to the full report or click on the Entrez Gene text at the upper left to return to Entrez Gene’s home page. The ‘Find related data’ menu in the column at the right allows selecting a database in which to find data related to initial query results. For example, to look for homologs of genes in a result set, select HomoloGene from the menu, read about how these links are calculated and click on ‘Find items’.