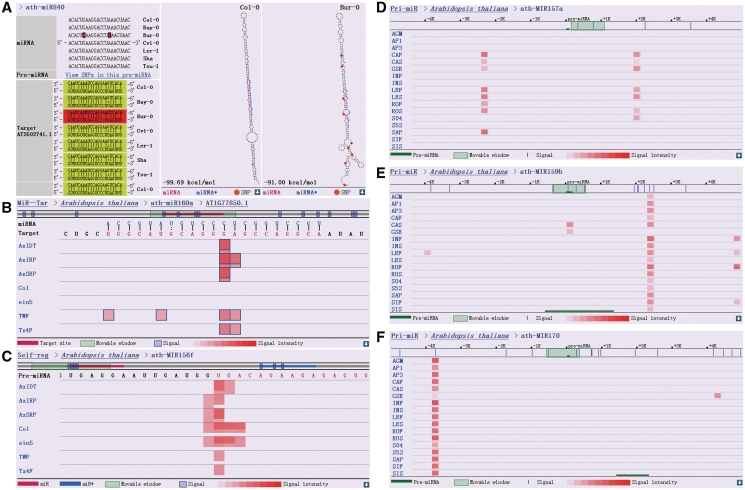
Figure 2.
Output data from the four major modules of PmiRKB. (A) An example of SNPs identified in the ‘SNP’ module (top left panel, highlighted in dark pink), which have the potential to influence the stability of miRNA–target RNA duplexes (lower left panel, indicated by different background colors). The effect of SNPs (indicated by orange circles) on the secondary structure of a specific pre-miRNA (precursor miRNA), ath-MIR840 for example, is illustrated on the right two panels. (B) Output data from the ‘MiR–Tar’ module that indicates AT1G77850.1 is a genuine target of ath-miR160a, based on the intense PARE signals detected in the middle of the miRNA–target duplex. The 100-nt sequence (gray horizontal line) surrounding the target site (red horizontal line) is shown. The blue outlines of the PARE signals denote the signatures that possess unique loci in the corresponding transcriptome. (C) The result given by ‘Self-reg’ indicates that the Dicer-like 1 (DCL1)-mediated first-step cleavages on the stem region of a specific pri-miRNA (primary miRNA) can be reflected by PARE data. In most cases, cleavage signals generated from the left arms of the pri-miRNAs were much easier to be detected than those from the right arms, likely due to the longer remnants generated from the left ones after DCL1-mediated cleavages. Cleavage signals within the miRNA* region of the pre-miRNA, ath-MIR156f, may represent miRNA-mediated self-regulation. For both (B and C), the signal intensity at a specific position is enhanced by the supporting of distinct signatures (represented by the red overlapping signals in different lengths in vertical orientation). (D, E and F) are output data from the ‘Pri-miR’ module. (D) represents tissue-specific transcription signals associated with the 3′-downstream region of ath-MIR157a, which could be hardly observed in the reproductive organs including inflorescences and siliques (the libraries lack of related signals, i.e. AGM, AP1, AP3, INF, INS, SIF, and SIS, were prepared from the reproductive organs of Arabidopsis). The transcription signals with distinct tissue-origin patterns were observed surrounding certain pre-miRNAs, such as ath-MIR159b (E) and ath-MIR170 (F), indicating that independent transcription units may exist. For (D, E and F), the signal intensity at a specific position is enhanced by the supporting of distinct signatures (represented by the red overlapping signals in different lengths in horizontal orientation).