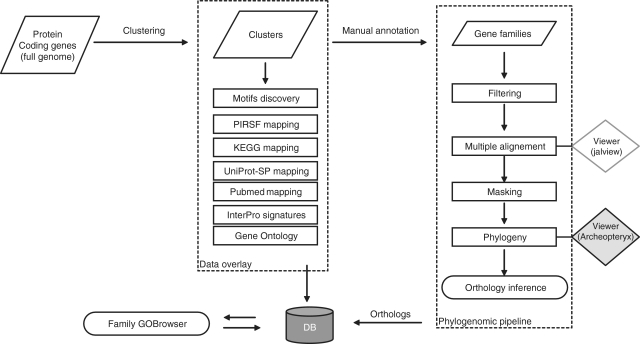
Figure 1.
Flowchart of the GreenPhylDB analyses. The input file is a multi-fasta file containing complete plant proteomes. In a first step, an automatic clustering aggregates all proteins in previously defined families. Sequences are classified as orphans if they cannot be regrouped in a cluster. Sequences composing the clusters are analyzed in order to overlay clusters with cross-references (e.g. UniProtKB, Pubmed, InterPro, MEME motifs, KEGG pathways data). Based on this information, clusters are manually curated in order to identify gene families. Finally, gene family sequences are analyzed via a phylogenetic-based pipeline to infer ortholog relationships. The procedure can be iterated for each new released genome using a lighter procedure. This ensures a cumulative and safe growth of the database. The data are stored in the database and can be easily accessed using dedicated visualizing tools including a gene tree viewer, a gene family browser and ortholog extracting tools.